| Full name: ankyrin repeat domain 45 | Alias Symbol: FLJ45235|CT117 | ||
| Type: protein-coding gene | Cytoband: 1q25.1 | ||
| Entrez ID: 339416 | HGNC ID: HGNC:24786 | Ensembl Gene: ENSG00000183831 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of ANKRD45:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ANKRD45 | 339416 | 236421_at | 0.0148 | 0.9466 | |
| GSE26886 | ANKRD45 | 339416 | 236421_at | 0.2469 | 0.0248 | |
| GSE45670 | ANKRD45 | 339416 | 236421_at | 0.0956 | 0.2374 | |
| GSE63941 | ANKRD45 | 339416 | 236421_at | -0.0966 | 0.4985 | |
| GSE77861 | ANKRD45 | 339416 | 236421_at | -0.0413 | 0.6004 | |
| GSE97050 | ANKRD45 | 339416 | A_33_P3219697 | -0.2212 | 0.3128 | |
| SRP133303 | ANKRD45 | 339416 | RNAseq | 0.3050 | 0.2020 | |
| TCGA | ANKRD45 | 339416 | RNAseq | -0.7388 | 0.1730 |
Upregulated datasets: 0; Downregulated datasets: 0.
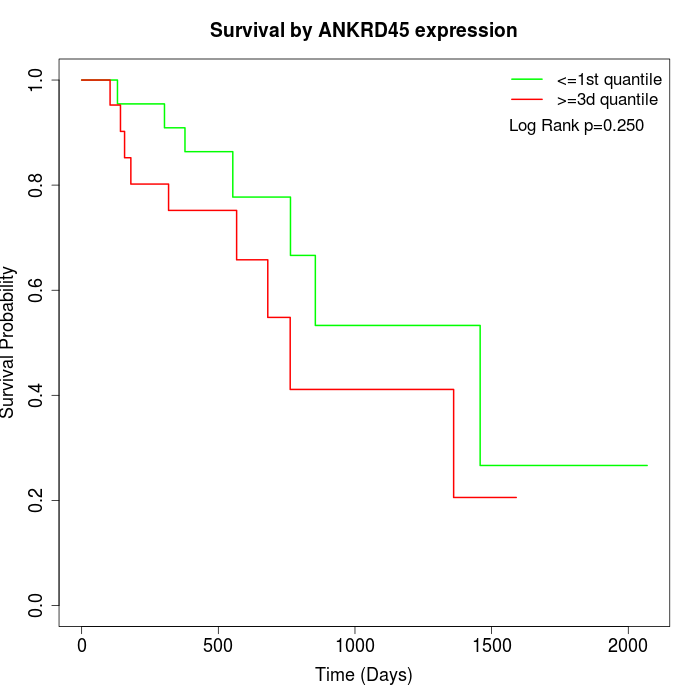
Survival by ANKRD45 expression:
Note: Click image to view full size file.
Copy number change of ANKRD45:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ANKRD45 | 339416 | 12 | 0 | 18 | |
| GSE20123 | ANKRD45 | 339416 | 12 | 0 | 18 | |
| GSE43470 | ANKRD45 | 339416 | 7 | 2 | 34 | |
| GSE46452 | ANKRD45 | 339416 | 3 | 1 | 55 | |
| GSE47630 | ANKRD45 | 339416 | 14 | 0 | 26 | |
| GSE54993 | ANKRD45 | 339416 | 0 | 6 | 64 | |
| GSE54994 | ANKRD45 | 339416 | 15 | 0 | 38 | |
| GSE60625 | ANKRD45 | 339416 | 0 | 0 | 11 | |
| GSE74703 | ANKRD45 | 339416 | 7 | 2 | 27 | |
| GSE74704 | ANKRD45 | 339416 | 5 | 0 | 15 | |
| TCGA | ANKRD45 | 339416 | 42 | 3 | 51 |
Total number of gains: 117; Total number of losses: 14; Total Number of normals: 357.
Somatic mutations of ANKRD45:
Generating mutation plots.
Highly correlated genes for ANKRD45:
Showing top 20/25 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ANKRD45 | RRP1 | 0.721385 | 3 | 0 | 3 |
| ANKRD45 | TNNI2 | 0.715029 | 3 | 0 | 3 |
| ANKRD45 | SLCO1A2 | 0.696847 | 3 | 0 | 3 |
| ANKRD45 | MAP7D2 | 0.689733 | 3 | 0 | 3 |
| ANKRD45 | ITGA2B | 0.677349 | 3 | 0 | 3 |
| ANKRD45 | APBA2 | 0.674701 | 3 | 0 | 3 |
| ANKRD45 | TTC25 | 0.657927 | 3 | 0 | 3 |
| ANKRD45 | ZBTB45 | 0.637398 | 4 | 0 | 4 |
| ANKRD45 | SUPT6H | 0.634763 | 3 | 0 | 3 |
| ANKRD45 | ELFN1-AS1 | 0.634429 | 3 | 0 | 3 |
| ANKRD45 | MAT1A | 0.613008 | 4 | 0 | 4 |
| ANKRD45 | FAM189B | 0.610912 | 4 | 0 | 3 |
| ANKRD45 | TNFAIP8L1 | 0.610766 | 4 | 0 | 3 |
| ANKRD45 | POR | 0.602055 | 3 | 0 | 3 |
| ANKRD45 | WDR90 | 0.601529 | 4 | 0 | 3 |
| ANKRD45 | FBRSL1 | 0.589283 | 4 | 0 | 3 |
| ANKRD45 | NAT14 | 0.583964 | 4 | 0 | 3 |
| ANKRD45 | SLC22A16 | 0.578139 | 3 | 0 | 3 |
| ANKRD45 | TNNI1 | 0.568152 | 4 | 0 | 3 |
| ANKRD45 | POLD1 | 0.564191 | 4 | 0 | 3 |
For details and further investigation, click here