| Full name: adaptor related protein complex 3 subunit sigma 2 | Alias Symbol: sigma3b | ||
| Type: protein-coding gene | Cytoband: 15q26.1 | ||
| Entrez ID: 10239 | HGNC ID: HGNC:571 | Ensembl Gene: ENSG00000157823 | OMIM ID: 602416 |
| Drug and gene relationship at DGIdb | |||
Expression of AP3S2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | AP3S2 | 10239 | 52482 | -0.4130 | 0.0009 | |
| GSE53624 | AP3S2 | 10239 | 12878 | -0.2674 | 0.0085 | |
| GSE97050 | AP3S2 | 10239 | A_24_P287691 | -0.1744 | 0.6347 | |
| SRP007169 | AP3S2 | 10239 | RNAseq | 0.1757 | 0.7033 | |
| SRP008496 | AP3S2 | 10239 | RNAseq | 0.1861 | 0.6911 | |
| SRP064894 | AP3S2 | 10239 | RNAseq | 0.1706 | 0.2210 | |
| SRP133303 | AP3S2 | 10239 | RNAseq | 0.4042 | 0.0363 | |
| SRP159526 | AP3S2 | 10239 | RNAseq | 0.4593 | 0.1833 | |
| SRP193095 | AP3S2 | 10239 | RNAseq | 0.2211 | 0.0441 | |
| SRP219564 | AP3S2 | 10239 | RNAseq | 0.2819 | 0.4901 | |
| TCGA | AP3S2 | 10239 | RNAseq | -0.1547 | 0.0013 |
Upregulated datasets: 0; Downregulated datasets: 0.
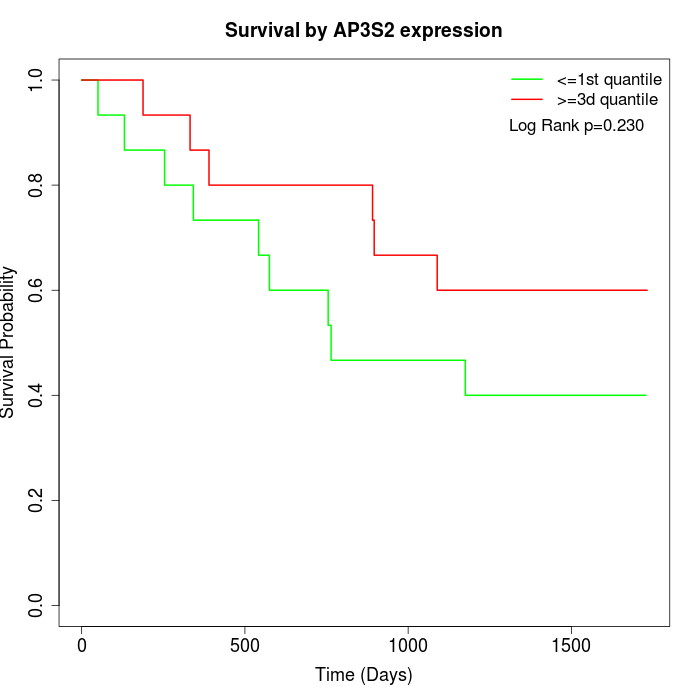
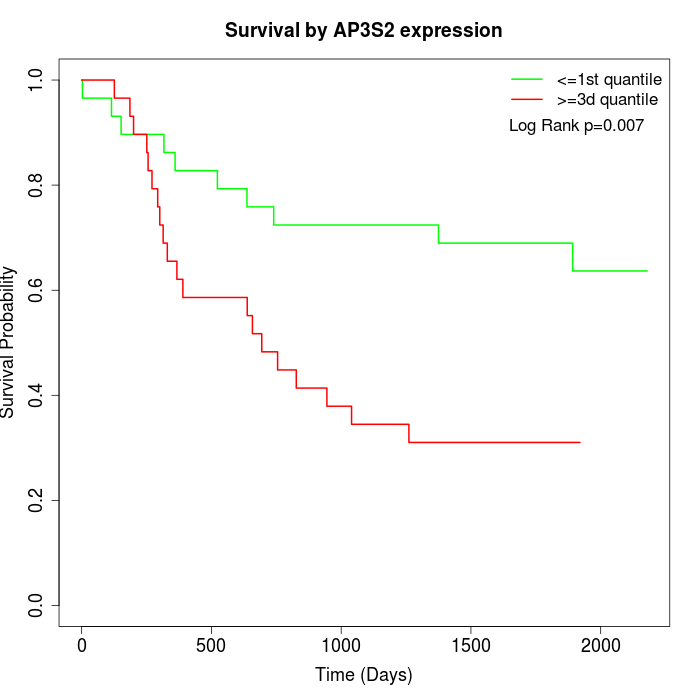
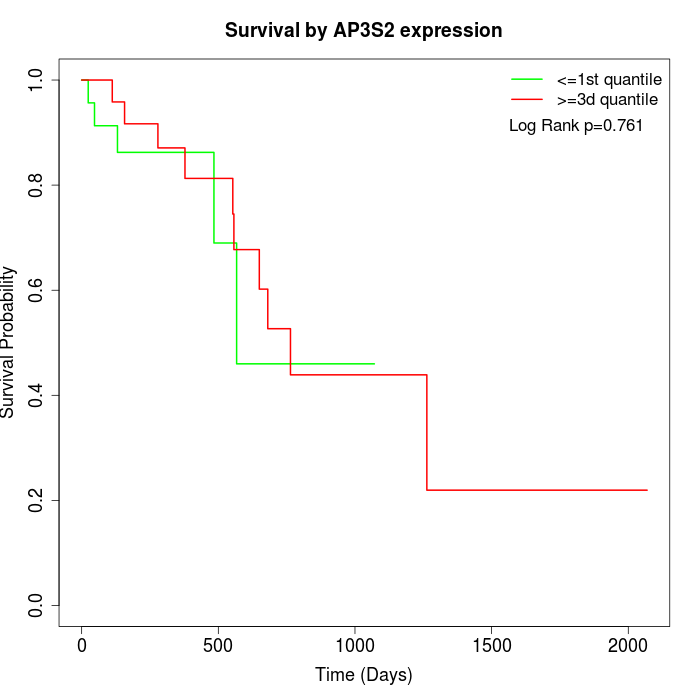
Survival by AP3S2 expression:
Note: Click image to view full size file.
Copy number change of AP3S2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | AP3S2 | 10239 | 8 | 3 | 19 | |
| GSE20123 | AP3S2 | 10239 | 8 | 3 | 19 | |
| GSE43470 | AP3S2 | 10239 | 5 | 4 | 34 | |
| GSE46452 | AP3S2 | 10239 | 3 | 7 | 49 | |
| GSE47630 | AP3S2 | 10239 | 8 | 11 | 21 | |
| GSE54993 | AP3S2 | 10239 | 4 | 6 | 60 | |
| GSE54994 | AP3S2 | 10239 | 7 | 6 | 40 | |
| GSE60625 | AP3S2 | 10239 | 4 | 0 | 7 | |
| GSE74703 | AP3S2 | 10239 | 4 | 3 | 29 | |
| GSE74704 | AP3S2 | 10239 | 4 | 2 | 14 | |
| TCGA | AP3S2 | 10239 | 18 | 12 | 66 |
Total number of gains: 73; Total number of losses: 57; Total Number of normals: 358.
Somatic mutations of AP3S2:
Generating mutation plots.
Highly correlated genes for AP3S2:
Showing top 20/86 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AP3S2 | TADA2B | 0.737888 | 3 | 0 | 3 |
| AP3S2 | DNAJC8 | 0.73777 | 3 | 0 | 3 |
| AP3S2 | BTD | 0.734822 | 3 | 0 | 3 |
| AP3S2 | ZNF510 | 0.730801 | 3 | 0 | 3 |
| AP3S2 | MYCT1 | 0.724901 | 3 | 0 | 3 |
| AP3S2 | STAT5A | 0.719851 | 3 | 0 | 3 |
| AP3S2 | CSTF2T | 0.719776 | 3 | 0 | 3 |
| AP3S2 | THAP2 | 0.71754 | 3 | 0 | 3 |
| AP3S2 | MED17 | 0.711536 | 3 | 0 | 3 |
| AP3S2 | IGIP | 0.710475 | 3 | 0 | 3 |
| AP3S2 | GOSR1 | 0.710339 | 3 | 0 | 3 |
| AP3S2 | APPBP2 | 0.7086 | 3 | 0 | 3 |
| AP3S2 | KLHL20 | 0.708486 | 3 | 0 | 3 |
| AP3S2 | SYNJ1 | 0.705724 | 3 | 0 | 3 |
| AP3S2 | CYYR1 | 0.703384 | 3 | 0 | 3 |
| AP3S2 | PLCL1 | 0.702148 | 3 | 0 | 3 |
| AP3S2 | PTPRB | 0.701076 | 3 | 0 | 3 |
| AP3S2 | RILP | 0.700851 | 3 | 0 | 3 |
| AP3S2 | SENP8 | 0.700034 | 3 | 0 | 3 |
| AP3S2 | MOCS2 | 0.699688 | 3 | 0 | 3 |
For details and further investigation, click here