| Full name: Rho/Rac guanine nucleotide exchange factor 2 | Alias Symbol: LFP40|GEF-H1|KIAA0651|P40|Lfc|GEFH1 | ||
| Type: protein-coding gene | Cytoband: 1q22 | ||
| Entrez ID: 9181 | HGNC ID: HGNC:682 | Ensembl Gene: ENSG00000116584 | OMIM ID: 607560 |
| Drug and gene relationship at DGIdb | |||
ARHGEF2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05130 | Pathogenic Escherichia coli infection |
Expression of ARHGEF2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ARHGEF2 | 9181 | 209435_s_at | 0.3468 | 0.2432 | |
| GSE20347 | ARHGEF2 | 9181 | 209435_s_at | 0.2915 | 0.0496 | |
| GSE23400 | ARHGEF2 | 9181 | 209435_s_at | 0.0712 | 0.1696 | |
| GSE26886 | ARHGEF2 | 9181 | 209435_s_at | -0.3106 | 0.2240 | |
| GSE29001 | ARHGEF2 | 9181 | 209435_s_at | 0.3409 | 0.2424 | |
| GSE38129 | ARHGEF2 | 9181 | 209435_s_at | 0.0444 | 0.8223 | |
| GSE45670 | ARHGEF2 | 9181 | 209435_s_at | 0.0462 | 0.8404 | |
| GSE53622 | ARHGEF2 | 9181 | 60785 | 0.1687 | 0.0542 | |
| GSE53624 | ARHGEF2 | 9181 | 60785 | 0.1944 | 0.0124 | |
| GSE63941 | ARHGEF2 | 9181 | 209435_s_at | 0.2325 | 0.7765 | |
| GSE77861 | ARHGEF2 | 9181 | 209435_s_at | 0.3393 | 0.0887 | |
| GSE97050 | ARHGEF2 | 9181 | A_23_P51699 | -0.3839 | 0.1496 | |
| SRP007169 | ARHGEF2 | 9181 | RNAseq | 0.3140 | 0.5077 | |
| SRP008496 | ARHGEF2 | 9181 | RNAseq | 0.1409 | 0.6762 | |
| SRP064894 | ARHGEF2 | 9181 | RNAseq | 0.4433 | 0.0287 | |
| SRP133303 | ARHGEF2 | 9181 | RNAseq | -0.0057 | 0.9738 | |
| SRP159526 | ARHGEF2 | 9181 | RNAseq | 0.6369 | 0.1440 | |
| SRP193095 | ARHGEF2 | 9181 | RNAseq | 0.3628 | 0.0734 | |
| SRP219564 | ARHGEF2 | 9181 | RNAseq | 0.3090 | 0.3034 | |
| TCGA | ARHGEF2 | 9181 | RNAseq | -0.1215 | 0.0239 |
Upregulated datasets: 0; Downregulated datasets: 0.
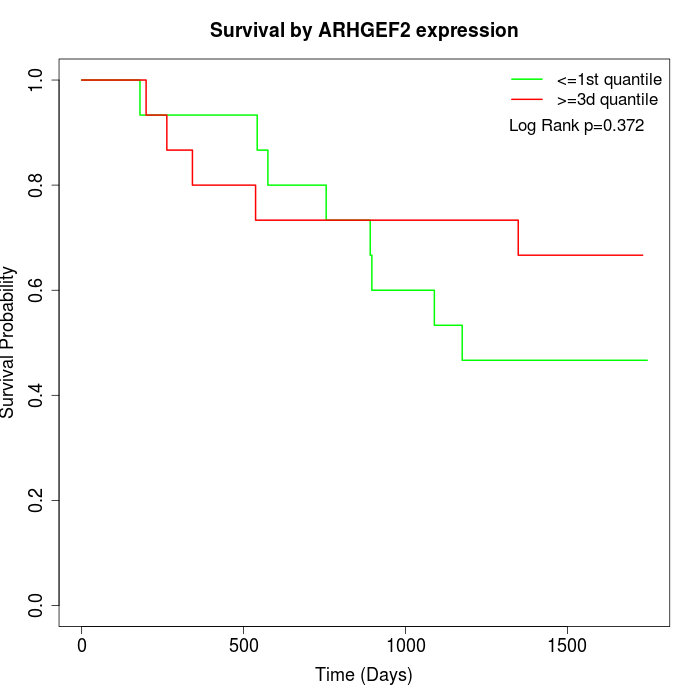
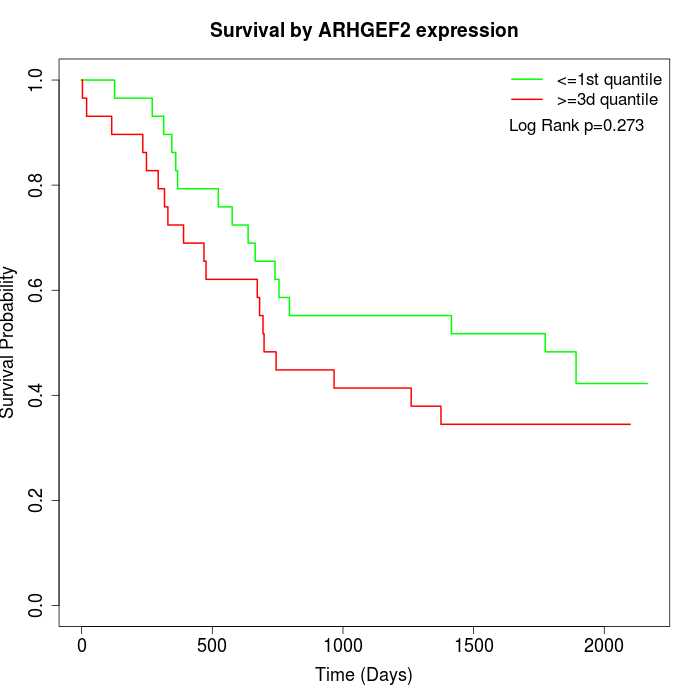
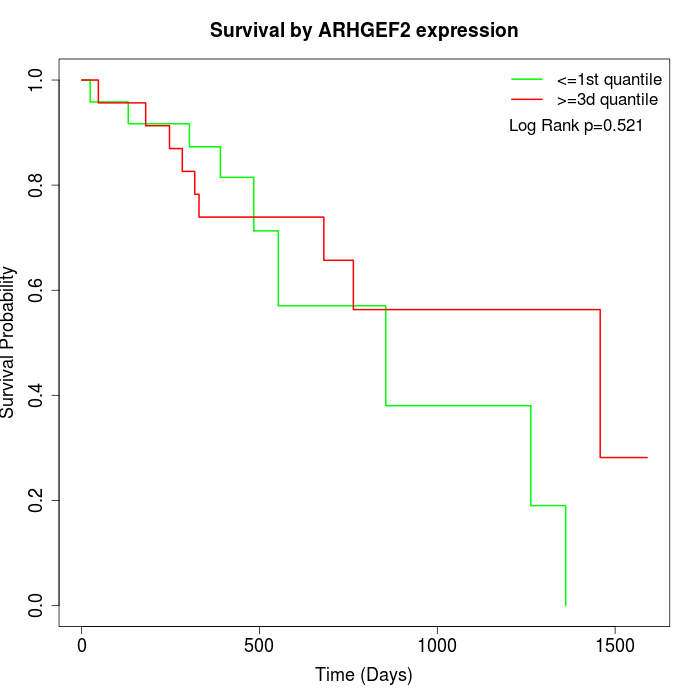
Survival by ARHGEF2 expression:
Note: Click image to view full size file.
Copy number change of ARHGEF2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ARHGEF2 | 9181 | 14 | 0 | 16 | |
| GSE20123 | ARHGEF2 | 9181 | 13 | 0 | 17 | |
| GSE43470 | ARHGEF2 | 9181 | 7 | 2 | 34 | |
| GSE46452 | ARHGEF2 | 9181 | 2 | 1 | 56 | |
| GSE47630 | ARHGEF2 | 9181 | 14 | 0 | 26 | |
| GSE54993 | ARHGEF2 | 9181 | 0 | 5 | 65 | |
| GSE54994 | ARHGEF2 | 9181 | 16 | 0 | 37 | |
| GSE60625 | ARHGEF2 | 9181 | 0 | 0 | 11 | |
| GSE74703 | ARHGEF2 | 9181 | 7 | 2 | 27 | |
| GSE74704 | ARHGEF2 | 9181 | 6 | 0 | 14 | |
| TCGA | ARHGEF2 | 9181 | 38 | 2 | 56 |
Total number of gains: 117; Total number of losses: 12; Total Number of normals: 359.
Somatic mutations of ARHGEF2:
Generating mutation plots.
Highly correlated genes for ARHGEF2:
Showing top 20/537 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ARHGEF2 | HELZ | 0.816621 | 3 | 0 | 3 |
| ARHGEF2 | FLOT2 | 0.740175 | 3 | 0 | 3 |
| ARHGEF2 | SPSB1 | 0.740028 | 3 | 0 | 3 |
| ARHGEF2 | SAMD1 | 0.738525 | 3 | 0 | 3 |
| ARHGEF2 | DDX59 | 0.728221 | 3 | 0 | 3 |
| ARHGEF2 | ARID1A | 0.721067 | 3 | 0 | 3 |
| ARHGEF2 | AAK1 | 0.718704 | 4 | 0 | 4 |
| ARHGEF2 | MED15 | 0.717934 | 3 | 0 | 3 |
| ARHGEF2 | CEP70 | 0.716024 | 3 | 0 | 3 |
| ARHGEF2 | RNF121 | 0.712782 | 3 | 0 | 3 |
| ARHGEF2 | ZNF764 | 0.71192 | 3 | 0 | 3 |
| ARHGEF2 | ZNF280C | 0.711049 | 4 | 0 | 3 |
| ARHGEF2 | SIX5 | 0.70759 | 4 | 0 | 3 |
| ARHGEF2 | USF2 | 0.701747 | 4 | 0 | 4 |
| ARHGEF2 | SLC39A1 | 0.699133 | 4 | 0 | 3 |
| ARHGEF2 | TMEM138 | 0.698541 | 3 | 0 | 3 |
| ARHGEF2 | NTPCR | 0.698297 | 3 | 0 | 3 |
| ARHGEF2 | C17orf67 | 0.697806 | 3 | 0 | 3 |
| ARHGEF2 | WDR5 | 0.696864 | 3 | 0 | 3 |
| ARHGEF2 | PPP1R9B | 0.691086 | 3 | 0 | 3 |
For details and further investigation, click here