| Full name: ATPase H+ transporting V0 subunit a1 | Alias Symbol: a1|Vph1|Stv1 | ||
| Type: protein-coding gene | Cytoband: 17q21.2 | ||
| Entrez ID: 535 | HGNC ID: HGNC:865 | Ensembl Gene: ENSG00000033627 | OMIM ID: 192130 |
| Drug and gene relationship at DGIdb | |||
ATP6V0A1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | |
| hsa05152 | Tuberculosis |
Expression of ATP6V0A1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V0A1 | 535 | 212383_at | -0.1794 | 0.6418 | |
| GSE20347 | ATP6V0A1 | 535 | 212383_at | -0.2589 | 0.0048 | |
| GSE23400 | ATP6V0A1 | 535 | 212383_at | -0.1985 | 0.0001 | |
| GSE26886 | ATP6V0A1 | 535 | 212383_at | -0.4221 | 0.0460 | |
| GSE29001 | ATP6V0A1 | 535 | 212383_at | -0.2510 | 0.3592 | |
| GSE38129 | ATP6V0A1 | 535 | 212383_at | -0.2446 | 0.0138 | |
| GSE45670 | ATP6V0A1 | 535 | 212383_at | 0.0761 | 0.5703 | |
| GSE53622 | ATP6V0A1 | 535 | 19396 | -0.5088 | 0.0000 | |
| GSE53624 | ATP6V0A1 | 535 | 19396 | -0.2488 | 0.0000 | |
| GSE63941 | ATP6V0A1 | 535 | 212383_at | 0.3001 | 0.4215 | |
| GSE77861 | ATP6V0A1 | 535 | 212383_at | -0.0473 | 0.8209 | |
| GSE97050 | ATP6V0A1 | 535 | A_24_P333733 | -0.0249 | 0.9295 | |
| SRP007169 | ATP6V0A1 | 535 | RNAseq | -0.4529 | 0.2069 | |
| SRP008496 | ATP6V0A1 | 535 | RNAseq | -0.6710 | 0.0021 | |
| SRP064894 | ATP6V0A1 | 535 | RNAseq | 0.1173 | 0.5190 | |
| SRP133303 | ATP6V0A1 | 535 | RNAseq | -0.1980 | 0.1297 | |
| SRP159526 | ATP6V0A1 | 535 | RNAseq | 0.1489 | 0.5690 | |
| SRP193095 | ATP6V0A1 | 535 | RNAseq | -0.0557 | 0.6053 | |
| SRP219564 | ATP6V0A1 | 535 | RNAseq | -0.1188 | 0.6517 | |
| TCGA | ATP6V0A1 | 535 | RNAseq | -0.1255 | 0.0161 |
Upregulated datasets: 0; Downregulated datasets: 0.
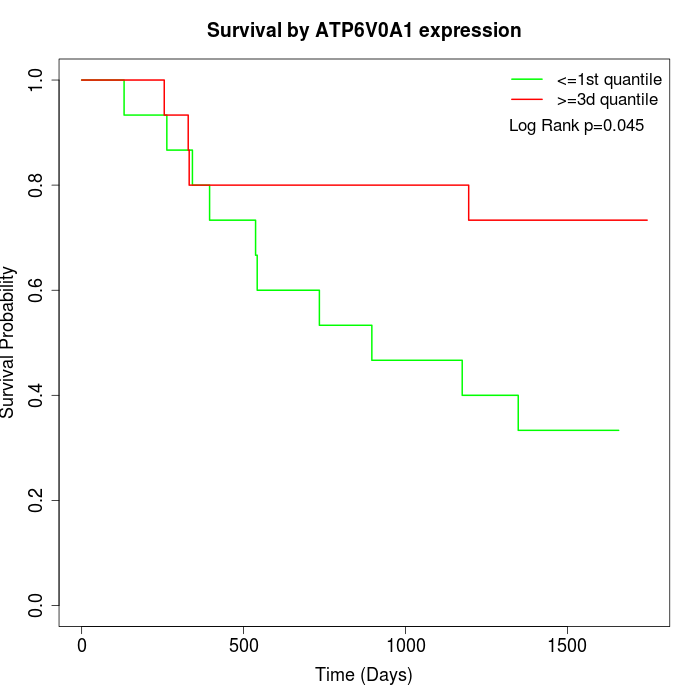
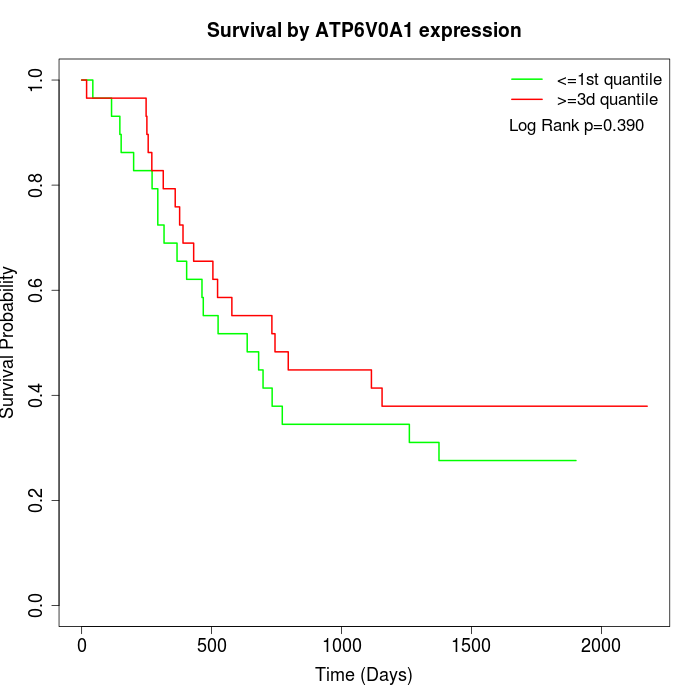
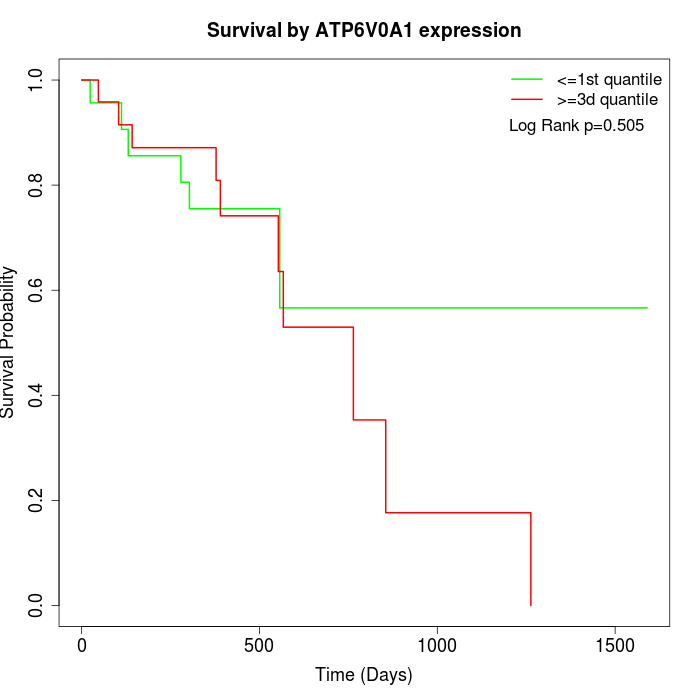
Survival by ATP6V0A1 expression:
Note: Click image to view full size file.
Copy number change of ATP6V0A1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V0A1 | 535 | 6 | 2 | 22 | |
| GSE20123 | ATP6V0A1 | 535 | 6 | 2 | 22 | |
| GSE43470 | ATP6V0A1 | 535 | 1 | 2 | 40 | |
| GSE46452 | ATP6V0A1 | 535 | 34 | 0 | 25 | |
| GSE47630 | ATP6V0A1 | 535 | 8 | 1 | 31 | |
| GSE54993 | ATP6V0A1 | 535 | 3 | 4 | 63 | |
| GSE54994 | ATP6V0A1 | 535 | 8 | 5 | 40 | |
| GSE60625 | ATP6V0A1 | 535 | 4 | 0 | 7 | |
| GSE74703 | ATP6V0A1 | 535 | 1 | 1 | 34 | |
| GSE74704 | ATP6V0A1 | 535 | 4 | 1 | 15 | |
| TCGA | ATP6V0A1 | 535 | 23 | 6 | 67 |
Total number of gains: 98; Total number of losses: 24; Total Number of normals: 366.
Somatic mutations of ATP6V0A1:
Generating mutation plots.
Highly correlated genes for ATP6V0A1:
Showing top 20/75 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V0A1 | UQCRC1 | 0.6677 | 3 | 0 | 3 |
| ATP6V0A1 | OVGP1 | 0.643154 | 4 | 0 | 3 |
| ATP6V0A1 | BDH1 | 0.626376 | 3 | 0 | 3 |
| ATP6V0A1 | ZNF287 | 0.624312 | 3 | 0 | 3 |
| ATP6V0A1 | HTR1E | 0.618558 | 3 | 0 | 3 |
| ATP6V0A1 | ACAP3 | 0.610993 | 4 | 0 | 3 |
| ATP6V0A1 | IRF2BP1 | 0.607152 | 3 | 0 | 3 |
| ATP6V0A1 | ACSL6 | 0.600467 | 4 | 0 | 3 |
| ATP6V0A1 | HIP1R | 0.600006 | 3 | 0 | 3 |
| ATP6V0A1 | UCK1 | 0.59908 | 4 | 0 | 4 |
| ATP6V0A1 | YBX2 | 0.596606 | 4 | 0 | 3 |
| ATP6V0A1 | LMNA | 0.59413 | 3 | 0 | 3 |
| ATP6V0A1 | PNPLA2 | 0.591235 | 5 | 0 | 4 |
| ATP6V0A1 | MAML3 | 0.583981 | 5 | 0 | 3 |
| ATP6V0A1 | ASTN1 | 0.583829 | 3 | 0 | 3 |
| ATP6V0A1 | FGF10 | 0.576522 | 3 | 0 | 3 |
| ATP6V0A1 | SIRT3 | 0.571621 | 4 | 0 | 3 |
| ATP6V0A1 | FKBP8 | 0.566271 | 6 | 0 | 3 |
| ATP6V0A1 | UBE4B | 0.56614 | 4 | 0 | 3 |
| ATP6V0A1 | SNX11 | 0.563937 | 5 | 0 | 4 |
For details and further investigation, click here