| Full name: ATPase H+ transporting V1 subunit A | Alias Symbol: Vma1|VA68 | ||
| Type: protein-coding gene | Cytoband: 3q13.31 | ||
| Entrez ID: 523 | HGNC ID: HGNC:851 | Ensembl Gene: ENSG00000114573 | OMIM ID: 607027 |
| Drug and gene relationship at DGIdb | |||
ATP6V1A involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04150 | mTOR signaling pathway | |
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection |
Expression of ATP6V1A:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V1A | 523 | 201972_at | 0.4876 | 0.3817 | |
| GSE20347 | ATP6V1A | 523 | 201972_at | -0.0635 | 0.6406 | |
| GSE23400 | ATP6V1A | 523 | 201972_at | 0.2794 | 0.0003 | |
| GSE26886 | ATP6V1A | 523 | 201972_at | -0.3637 | 0.0131 | |
| GSE29001 | ATP6V1A | 523 | 201972_at | 0.0296 | 0.9033 | |
| GSE38129 | ATP6V1A | 523 | 201972_at | 0.1041 | 0.5974 | |
| GSE45670 | ATP6V1A | 523 | 201972_at | 0.1156 | 0.3697 | |
| GSE53622 | ATP6V1A | 523 | 35986 | -0.0188 | 0.8430 | |
| GSE53624 | ATP6V1A | 523 | 35986 | -0.0949 | 0.1243 | |
| GSE63941 | ATP6V1A | 523 | 201972_at | -0.6748 | 0.0451 | |
| GSE77861 | ATP6V1A | 523 | 201972_at | -0.3618 | 0.3965 | |
| GSE97050 | ATP6V1A | 523 | A_24_P396994 | 0.5872 | 0.2054 | |
| SRP007169 | ATP6V1A | 523 | RNAseq | -0.7172 | 0.1633 | |
| SRP008496 | ATP6V1A | 523 | RNAseq | -0.2746 | 0.4204 | |
| SRP064894 | ATP6V1A | 523 | RNAseq | -0.0143 | 0.9501 | |
| SRP133303 | ATP6V1A | 523 | RNAseq | 0.1115 | 0.5990 | |
| SRP159526 | ATP6V1A | 523 | RNAseq | 0.0621 | 0.8092 | |
| SRP193095 | ATP6V1A | 523 | RNAseq | -0.0927 | 0.4790 | |
| SRP219564 | ATP6V1A | 523 | RNAseq | -0.1271 | 0.7928 | |
| TCGA | ATP6V1A | 523 | RNAseq | 0.0455 | 0.3910 |
Upregulated datasets: 0; Downregulated datasets: 0.
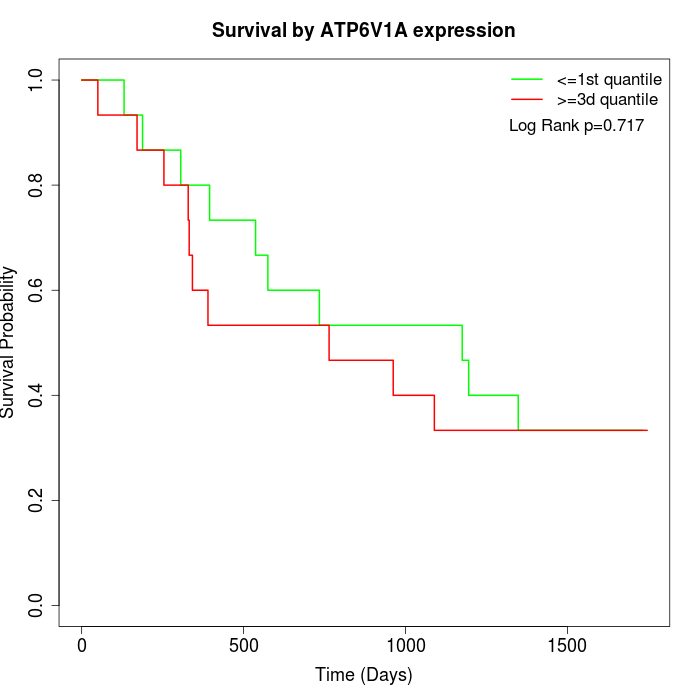
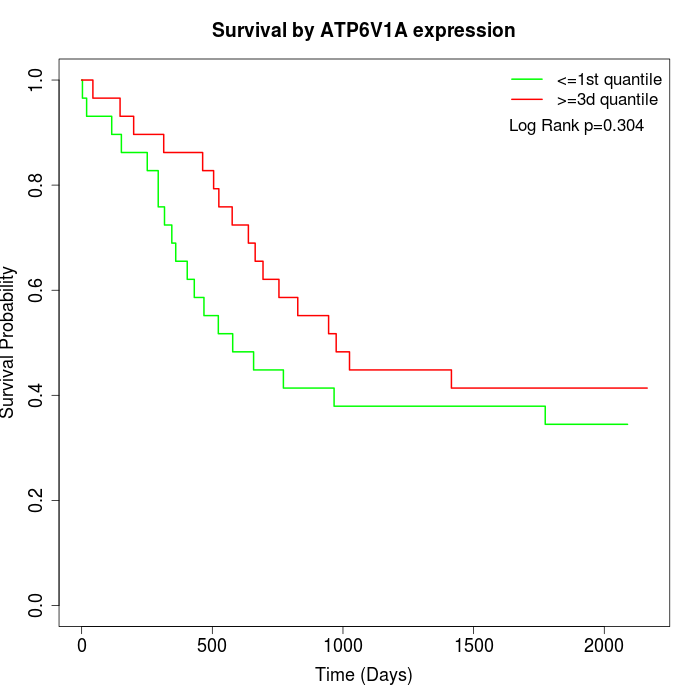
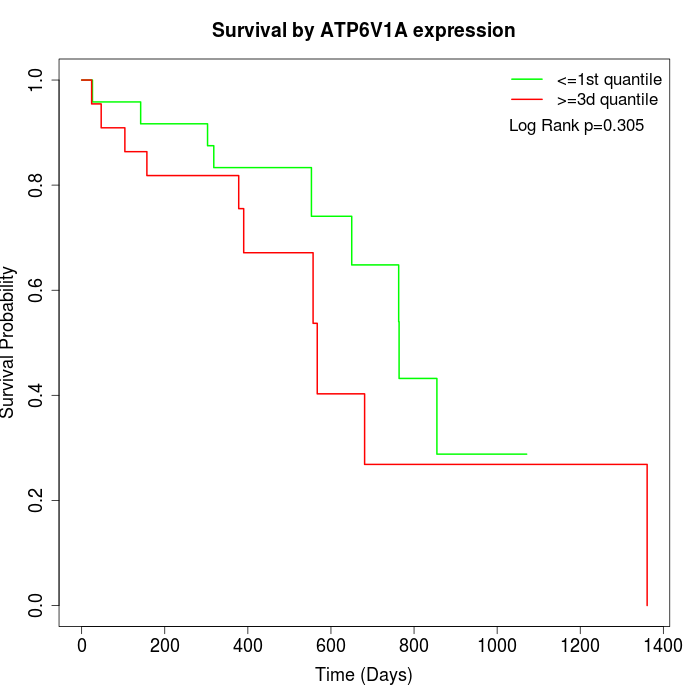
Survival by ATP6V1A expression:
Note: Click image to view full size file.
Copy number change of ATP6V1A:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V1A | 523 | 16 | 1 | 13 | |
| GSE20123 | ATP6V1A | 523 | 16 | 1 | 13 | |
| GSE43470 | ATP6V1A | 523 | 21 | 1 | 21 | |
| GSE46452 | ATP6V1A | 523 | 13 | 5 | 41 | |
| GSE47630 | ATP6V1A | 523 | 16 | 6 | 18 | |
| GSE54993 | ATP6V1A | 523 | 2 | 6 | 62 | |
| GSE54994 | ATP6V1A | 523 | 29 | 4 | 20 | |
| GSE60625 | ATP6V1A | 523 | 0 | 6 | 5 | |
| GSE74703 | ATP6V1A | 523 | 17 | 1 | 18 | |
| GSE74704 | ATP6V1A | 523 | 12 | 1 | 7 | |
| TCGA | ATP6V1A | 523 | 52 | 7 | 37 |
Total number of gains: 194; Total number of losses: 39; Total Number of normals: 255.
Somatic mutations of ATP6V1A:
Generating mutation plots.
Highly correlated genes for ATP6V1A:
Showing top 20/519 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V1A | PUSL1 | 0.744317 | 3 | 0 | 3 |
| ATP6V1A | ARHGEF18 | 0.736784 | 3 | 0 | 3 |
| ATP6V1A | LRP10 | 0.721522 | 3 | 0 | 3 |
| ATP6V1A | RXRA | 0.718852 | 4 | 0 | 4 |
| ATP6V1A | PPP4R2 | 0.713061 | 3 | 0 | 3 |
| ATP6V1A | WDR37 | 0.708845 | 3 | 0 | 3 |
| ATP6V1A | HDAC1 | 0.707534 | 4 | 0 | 3 |
| ATP6V1A | PRADC1 | 0.706131 | 3 | 0 | 3 |
| ATP6V1A | C1D | 0.704643 | 3 | 0 | 3 |
| ATP6V1A | TMEM117 | 0.704256 | 3 | 0 | 3 |
| ATP6V1A | RSPRY1 | 0.699465 | 3 | 0 | 3 |
| ATP6V1A | SYNRG | 0.698817 | 3 | 0 | 3 |
| ATP6V1A | TRIP12 | 0.69494 | 4 | 0 | 4 |
| ATP6V1A | RNF19B | 0.693469 | 3 | 0 | 3 |
| ATP6V1A | CKAP4 | 0.693225 | 3 | 0 | 3 |
| ATP6V1A | MAPK8 | 0.692751 | 3 | 0 | 3 |
| ATP6V1A | SMARCA4 | 0.686143 | 3 | 0 | 3 |
| ATP6V1A | TMEM167A | 0.685033 | 4 | 0 | 3 |
| ATP6V1A | NCK2 | 0.682689 | 3 | 0 | 3 |
| ATP6V1A | TMEM41B | 0.681756 | 4 | 0 | 4 |
For details and further investigation, click here