| Full name: thyroid hormone receptor interactor 12 | Alias Symbol: KIAA0045 | ||
| Type: protein-coding gene | Cytoband: 2q36.3 | ||
| Entrez ID: 9320 | HGNC ID: HGNC:12306 | Ensembl Gene: ENSG00000153827 | OMIM ID: 604506 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of TRIP12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIP12 | 9320 | 201546_at | -0.2866 | 0.4222 | |
| GSE20347 | TRIP12 | 9320 | 201546_at | -0.6010 | 0.0000 | |
| GSE23400 | TRIP12 | 9320 | 201546_at | -0.2265 | 0.0009 | |
| GSE26886 | TRIP12 | 9320 | 201546_at | -1.2696 | 0.0000 | |
| GSE29001 | TRIP12 | 9320 | 201546_at | -0.4906 | 0.0853 | |
| GSE38129 | TRIP12 | 9320 | 201546_at | -0.4143 | 0.0003 | |
| GSE45670 | TRIP12 | 9320 | 201546_at | 0.0870 | 0.4498 | |
| GSE53622 | TRIP12 | 9320 | 50174 | -0.3628 | 0.0000 | |
| GSE53624 | TRIP12 | 9320 | 50174 | -0.2324 | 0.0000 | |
| GSE63941 | TRIP12 | 9320 | 201546_at | -1.0303 | 0.0039 | |
| GSE77861 | TRIP12 | 9320 | 201546_at | -0.0859 | 0.7232 | |
| GSE97050 | TRIP12 | 9320 | A_23_P147984 | -0.1977 | 0.3956 | |
| SRP007169 | TRIP12 | 9320 | RNAseq | -0.4181 | 0.2224 | |
| SRP008496 | TRIP12 | 9320 | RNAseq | -0.2887 | 0.0818 | |
| SRP064894 | TRIP12 | 9320 | RNAseq | -0.2317 | 0.1517 | |
| SRP133303 | TRIP12 | 9320 | RNAseq | 0.0274 | 0.7428 | |
| SRP159526 | TRIP12 | 9320 | RNAseq | -0.4810 | 0.0216 | |
| SRP193095 | TRIP12 | 9320 | RNAseq | -0.3314 | 0.0000 | |
| SRP219564 | TRIP12 | 9320 | RNAseq | -0.4962 | 0.0586 | |
| TCGA | TRIP12 | 9320 | RNAseq | -0.0803 | 0.0540 |
Upregulated datasets: 0; Downregulated datasets: 2.
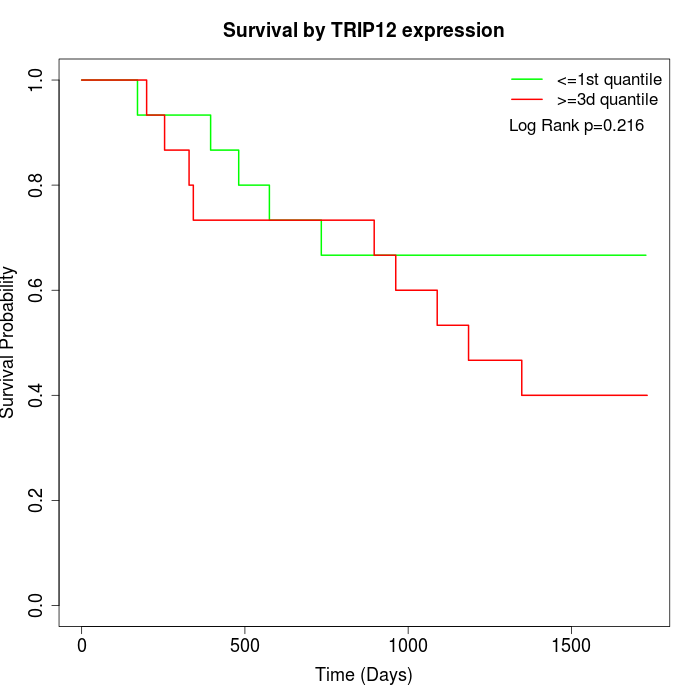
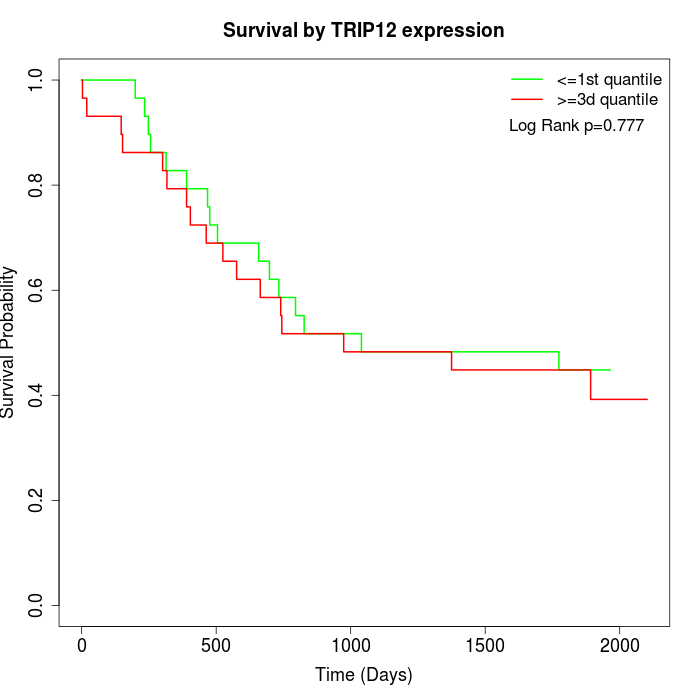
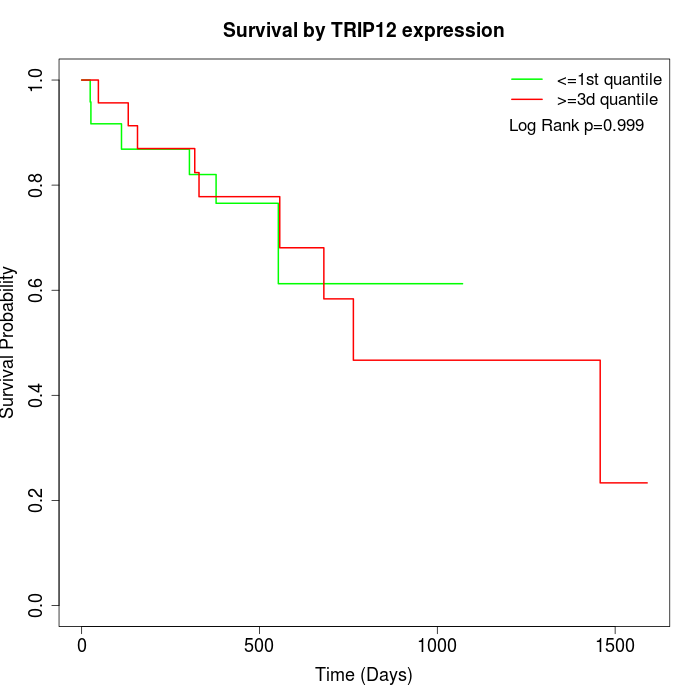
Survival by TRIP12 expression:
Note: Click image to view full size file.
Copy number change of TRIP12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIP12 | 9320 | 1 | 13 | 16 | |
| GSE20123 | TRIP12 | 9320 | 1 | 13 | 16 | |
| GSE43470 | TRIP12 | 9320 | 2 | 8 | 33 | |
| GSE46452 | TRIP12 | 9320 | 0 | 5 | 54 | |
| GSE47630 | TRIP12 | 9320 | 4 | 5 | 31 | |
| GSE54993 | TRIP12 | 9320 | 1 | 2 | 67 | |
| GSE54994 | TRIP12 | 9320 | 6 | 13 | 34 | |
| GSE60625 | TRIP12 | 9320 | 0 | 3 | 8 | |
| GSE74703 | TRIP12 | 9320 | 2 | 7 | 27 | |
| GSE74704 | TRIP12 | 9320 | 1 | 6 | 13 | |
| TCGA | TRIP12 | 9320 | 13 | 27 | 56 |
Total number of gains: 31; Total number of losses: 102; Total Number of normals: 355.
Somatic mutations of TRIP12:
Generating mutation plots.
Highly correlated genes for TRIP12:
Showing top 20/1229 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIP12 | CMTR2 | 0.818001 | 3 | 0 | 3 |
| TRIP12 | RAB21 | 0.81719 | 4 | 0 | 4 |
| TRIP12 | FYTTD1 | 0.777531 | 3 | 0 | 3 |
| TRIP12 | CAPNS2 | 0.766517 | 4 | 0 | 4 |
| TRIP12 | MRVI1 | 0.758923 | 3 | 0 | 3 |
| TRIP12 | NAA20 | 0.750073 | 3 | 0 | 3 |
| TRIP12 | MAN2B2 | 0.745364 | 3 | 0 | 3 |
| TRIP12 | ARPC1A | 0.742594 | 4 | 0 | 3 |
| TRIP12 | MAT2B | 0.742457 | 3 | 0 | 3 |
| TRIP12 | EPG5 | 0.735162 | 5 | 0 | 5 |
| TRIP12 | UBQLN1 | 0.730675 | 3 | 0 | 3 |
| TRIP12 | FAHD1 | 0.725844 | 4 | 0 | 3 |
| TRIP12 | STAU2 | 0.723478 | 5 | 0 | 4 |
| TRIP12 | CNEP1R1 | 0.721095 | 4 | 0 | 4 |
| TRIP12 | PPIL4 | 0.713272 | 3 | 0 | 3 |
| TRIP12 | ARHGEF6 | 0.70559 | 3 | 0 | 3 |
| TRIP12 | ITGB5 | 0.704967 | 3 | 0 | 3 |
| TRIP12 | CHL1 | 0.704432 | 3 | 0 | 3 |
| TRIP12 | GOLGA5 | 0.702815 | 3 | 0 | 3 |
| TRIP12 | PPP2R5E | 0.70173 | 4 | 0 | 3 |
For details and further investigation, click here