| Full name: BCL2 associated X, apoptosis regulator | Alias Symbol: BCL2L4 | ||
| Type: protein-coding gene | Cytoband: 19q13.33 | ||
| Entrez ID: 581 | HGNC ID: HGNC:959 | Ensembl Gene: ENSG00000087088 | OMIM ID: 600040 |
| Drug and gene relationship at DGIdb | |||
BAX involved pathways:
Expression of BAX:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BAX | 581 | 211833_s_at | 0.5236 | 0.0545 | |
| GSE20347 | BAX | 581 | 211833_s_at | 0.0546 | 0.7789 | |
| GSE23400 | BAX | 581 | 211833_s_at | 0.5652 | 0.0000 | |
| GSE26886 | BAX | 581 | 211833_s_at | -0.1210 | 0.4783 | |
| GSE29001 | BAX | 581 | 211833_s_at | 0.5309 | 0.2551 | |
| GSE38129 | BAX | 581 | 211833_s_at | 0.4852 | 0.1120 | |
| GSE45670 | BAX | 581 | 211833_s_at | 0.5212 | 0.0005 | |
| GSE53622 | BAX | 581 | 4054 | -0.0341 | 0.6711 | |
| GSE53624 | BAX | 581 | 4054 | 0.2039 | 0.0048 | |
| GSE63941 | BAX | 581 | 211833_s_at | -0.7296 | 0.0930 | |
| GSE77861 | BAX | 581 | 208478_s_at | 0.4843 | 0.0587 | |
| GSE97050 | BAX | 581 | A_23_P208706 | 0.2826 | 0.2776 | |
| SRP007169 | BAX | 581 | RNAseq | 0.3636 | 0.2678 | |
| SRP008496 | BAX | 581 | RNAseq | 0.2175 | 0.2946 | |
| SRP064894 | BAX | 581 | RNAseq | 1.2937 | 0.0000 | |
| SRP133303 | BAX | 581 | RNAseq | 0.4221 | 0.0156 | |
| SRP159526 | BAX | 581 | RNAseq | 0.6560 | 0.0006 | |
| SRP193095 | BAX | 581 | RNAseq | 0.2473 | 0.0440 | |
| SRP219564 | BAX | 581 | RNAseq | 0.4082 | 0.3992 | |
| TCGA | BAX | 581 | RNAseq | 0.3648 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
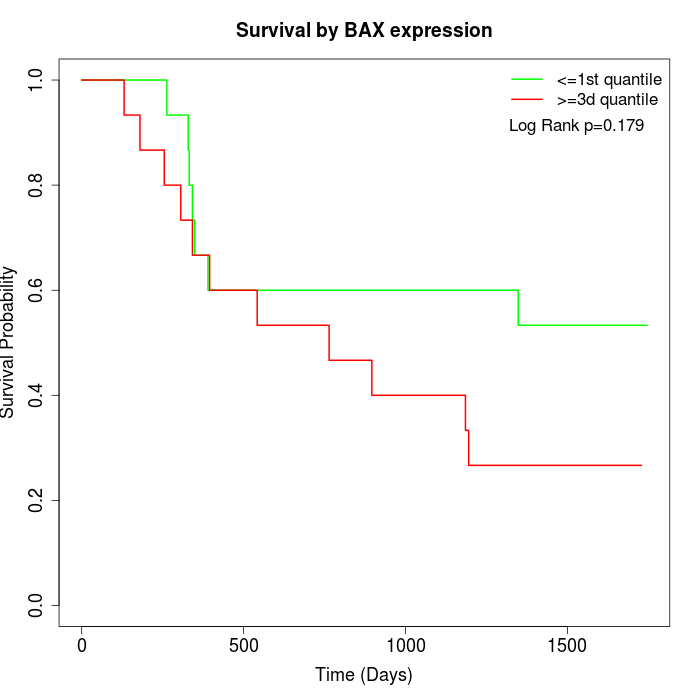
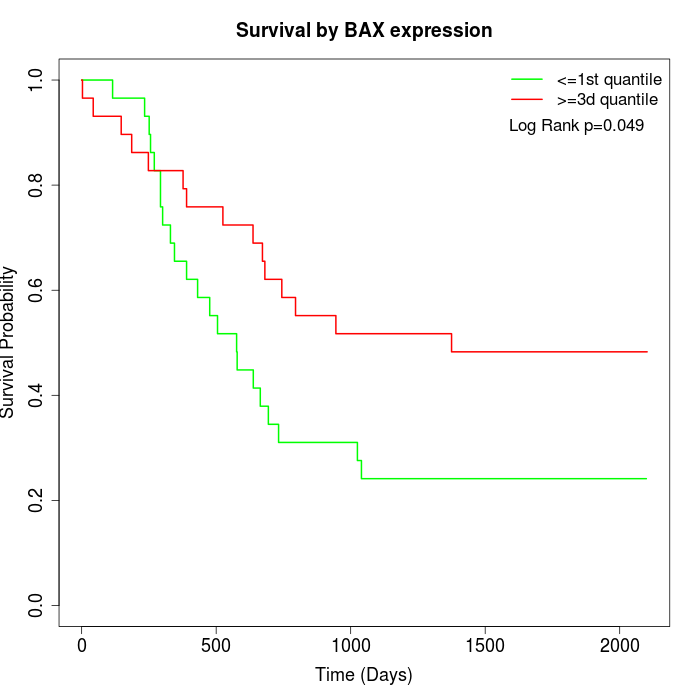
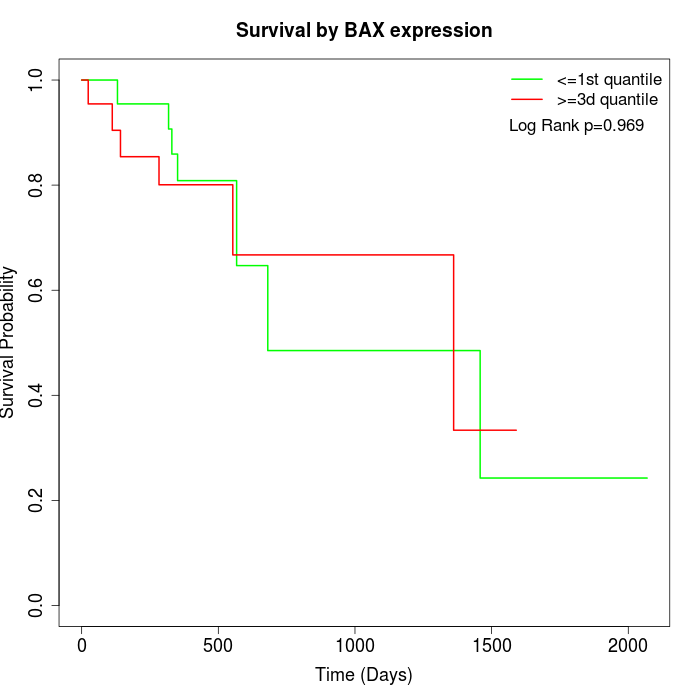
Survival by BAX expression:
Note: Click image to view full size file.
Copy number change of BAX:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BAX | 581 | 4 | 4 | 22 | |
| GSE20123 | BAX | 581 | 4 | 3 | 23 | |
| GSE43470 | BAX | 581 | 4 | 10 | 29 | |
| GSE46452 | BAX | 581 | 45 | 1 | 13 | |
| GSE47630 | BAX | 581 | 9 | 6 | 25 | |
| GSE54993 | BAX | 581 | 17 | 4 | 49 | |
| GSE54994 | BAX | 581 | 4 | 14 | 35 | |
| GSE60625 | BAX | 581 | 9 | 0 | 2 | |
| GSE74703 | BAX | 581 | 4 | 7 | 25 | |
| GSE74704 | BAX | 581 | 4 | 1 | 15 | |
| TCGA | BAX | 581 | 14 | 18 | 64 |
Total number of gains: 118; Total number of losses: 68; Total Number of normals: 302.
Somatic mutations of BAX:
Generating mutation plots.
Highly correlated genes for BAX:
Showing top 20/273 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BAX | POC1A | 0.803111 | 3 | 0 | 3 |
| BAX | CECR2 | 0.789332 | 3 | 0 | 3 |
| BAX | C4orf46 | 0.759275 | 3 | 0 | 3 |
| BAX | FUT1 | 0.73435 | 4 | 0 | 4 |
| BAX | TKTL1 | 0.73192 | 3 | 0 | 3 |
| BAX | TXNDC12 | 0.728073 | 3 | 0 | 3 |
| BAX | ATG4D | 0.716125 | 3 | 0 | 3 |
| BAX | CMSS1 | 0.715475 | 3 | 0 | 3 |
| BAX | MEX3D | 0.713661 | 6 | 0 | 5 |
| BAX | TRAIP | 0.712517 | 4 | 0 | 4 |
| BAX | UBE2S | 0.710062 | 8 | 0 | 8 |
| BAX | CHP2 | 0.709254 | 3 | 0 | 3 |
| BAX | MTERF3 | 0.708019 | 3 | 0 | 3 |
| BAX | RASGRF2 | 0.707226 | 3 | 0 | 3 |
| BAX | TRIM11 | 0.705042 | 4 | 0 | 3 |
| BAX | RAD54B | 0.702937 | 5 | 0 | 4 |
| BAX | YEATS2 | 0.697884 | 4 | 0 | 4 |
| BAX | ZNF687 | 0.695845 | 3 | 0 | 3 |
| BAX | WDR46 | 0.695034 | 7 | 0 | 6 |
| BAX | TMCC2 | 0.694607 | 4 | 0 | 3 |
For details and further investigation, click here