| Full name: BCAS3 microtubule associated cell migration factor | Alias Symbol: FLJ20128 | ||
| Type: protein-coding gene | Cytoband: 17q23.2 | ||
| Entrez ID: 54828 | HGNC ID: HGNC:14347 | Ensembl Gene: ENSG00000141376 | OMIM ID: 607470 |
| Drug and gene relationship at DGIdb | |||
Expression of BCAS3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BCAS3 | 54828 | 220488_s_at | -0.0538 | 0.9044 | |
| GSE20347 | BCAS3 | 54828 | 220488_s_at | -0.0259 | 0.8250 | |
| GSE23400 | BCAS3 | 54828 | 220488_s_at | -0.0828 | 0.0722 | |
| GSE26886 | BCAS3 | 54828 | 220488_s_at | 0.7392 | 0.0235 | |
| GSE29001 | BCAS3 | 54828 | 220488_s_at | -0.1388 | 0.5085 | |
| GSE38129 | BCAS3 | 54828 | 220488_s_at | 0.0964 | 0.4362 | |
| GSE45670 | BCAS3 | 54828 | 220488_s_at | 0.0084 | 0.9666 | |
| GSE53622 | BCAS3 | 54828 | 17194 | 0.1573 | 0.0264 | |
| GSE53624 | BCAS3 | 54828 | 33445 | -0.0113 | 0.9024 | |
| GSE63941 | BCAS3 | 54828 | 220488_s_at | 0.0319 | 0.9545 | |
| GSE77861 | BCAS3 | 54828 | 220488_s_at | 0.2591 | 0.3780 | |
| GSE97050 | BCAS3 | 54828 | A_24_P100761 | -0.2688 | 0.2401 | |
| SRP007169 | BCAS3 | 54828 | RNAseq | 0.6338 | 0.1985 | |
| SRP008496 | BCAS3 | 54828 | RNAseq | 0.6698 | 0.0411 | |
| SRP064894 | BCAS3 | 54828 | RNAseq | 0.2214 | 0.1731 | |
| SRP133303 | BCAS3 | 54828 | RNAseq | 0.1843 | 0.1313 | |
| SRP159526 | BCAS3 | 54828 | RNAseq | 0.4249 | 0.2264 | |
| SRP193095 | BCAS3 | 54828 | RNAseq | 0.1188 | 0.2209 | |
| SRP219564 | BCAS3 | 54828 | RNAseq | -0.1589 | 0.5599 | |
| TCGA | BCAS3 | 54828 | RNAseq | 0.0654 | 0.2924 |
Upregulated datasets: 0; Downregulated datasets: 0.
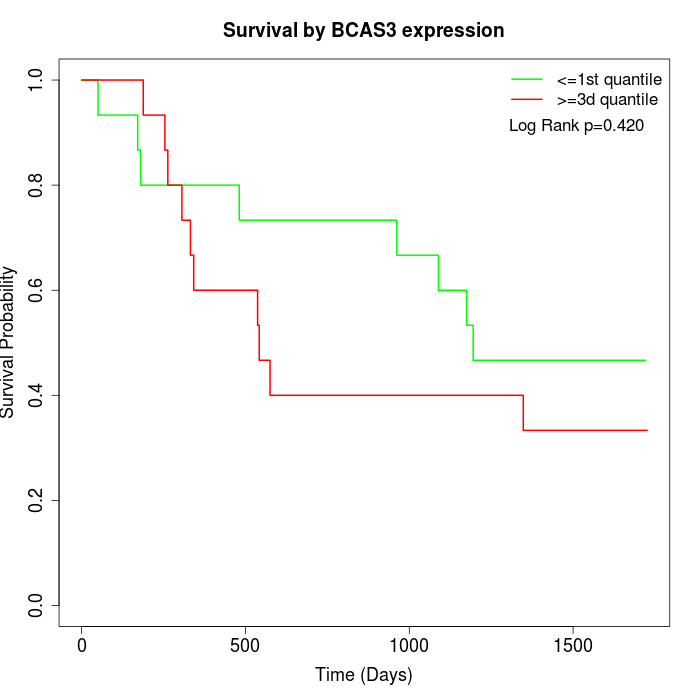
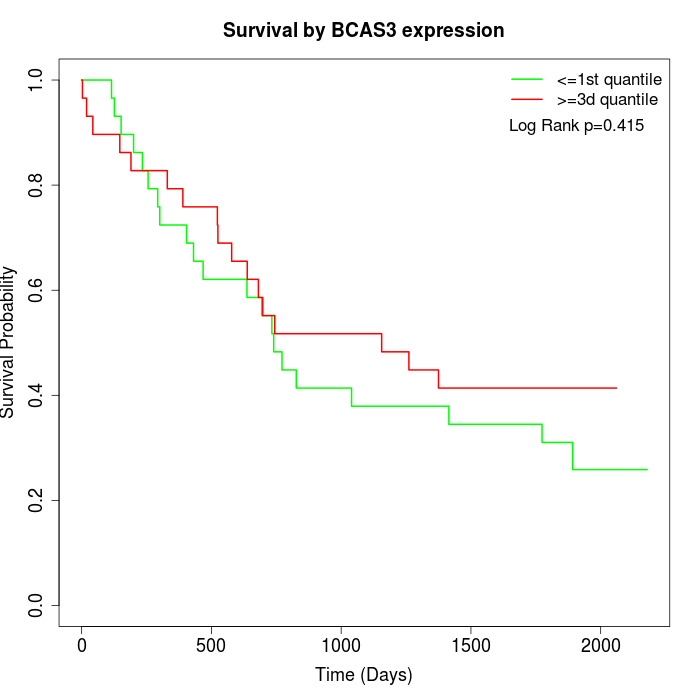
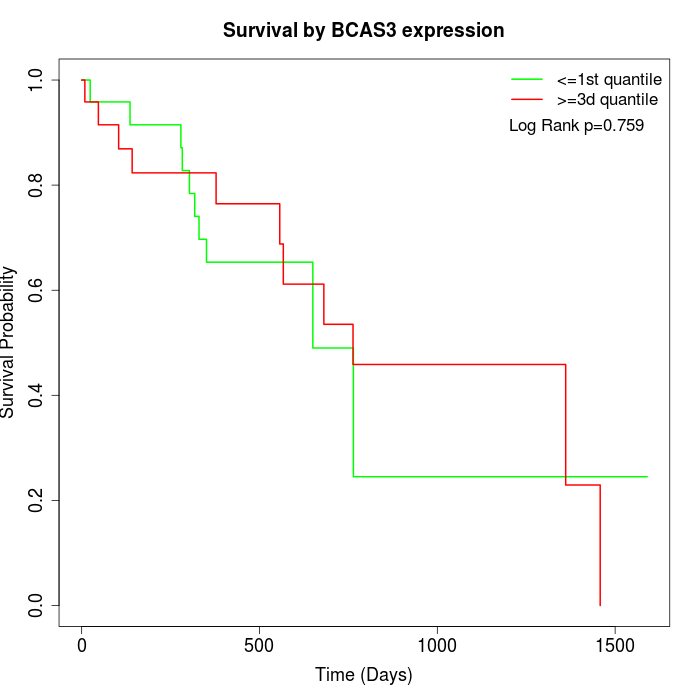
Survival by BCAS3 expression:
Note: Click image to view full size file.
Copy number change of BCAS3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BCAS3 | 54828 | 5 | 1 | 24 | |
| GSE20123 | BCAS3 | 54828 | 5 | 1 | 24 | |
| GSE43470 | BCAS3 | 54828 | 5 | 0 | 38 | |
| GSE46452 | BCAS3 | 54828 | 32 | 0 | 27 | |
| GSE47630 | BCAS3 | 54828 | 7 | 1 | 32 | |
| GSE54993 | BCAS3 | 54828 | 2 | 5 | 63 | |
| GSE54994 | BCAS3 | 54828 | 9 | 5 | 39 | |
| GSE60625 | BCAS3 | 54828 | 4 | 0 | 7 | |
| GSE74703 | BCAS3 | 54828 | 5 | 0 | 31 | |
| GSE74704 | BCAS3 | 54828 | 4 | 1 | 15 | |
| TCGA | BCAS3 | 54828 | 31 | 8 | 57 |
Total number of gains: 109; Total number of losses: 22; Total Number of normals: 357.
Somatic mutations of BCAS3:
Generating mutation plots.
Highly correlated genes for BCAS3:
Showing top 20/305 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BCAS3 | WDR35 | 0.745214 | 3 | 0 | 3 |
| BCAS3 | ZMAT1 | 0.723628 | 3 | 0 | 3 |
| BCAS3 | HSD17B1 | 0.717598 | 3 | 0 | 3 |
| BCAS3 | MAP1LC3A | 0.703551 | 3 | 0 | 3 |
| BCAS3 | GABARAP | 0.702142 | 3 | 0 | 3 |
| BCAS3 | PFKM | 0.696255 | 3 | 0 | 3 |
| BCAS3 | CAT | 0.692188 | 4 | 0 | 3 |
| BCAS3 | STAG2 | 0.690061 | 3 | 0 | 3 |
| BCAS3 | ZNHIT3 | 0.687266 | 4 | 0 | 4 |
| BCAS3 | CENPV | 0.680659 | 3 | 0 | 3 |
| BCAS3 | R3HDM2 | 0.67447 | 3 | 0 | 3 |
| BCAS3 | CTSO | 0.673973 | 4 | 0 | 3 |
| BCAS3 | BSDC1 | 0.673325 | 3 | 0 | 3 |
| BCAS3 | TCF21 | 0.673302 | 3 | 0 | 3 |
| BCAS3 | SKP1 | 0.667368 | 3 | 0 | 3 |
| BCAS3 | LMO1 | 0.666993 | 3 | 0 | 3 |
| BCAS3 | CREB3 | 0.66648 | 4 | 0 | 3 |
| BCAS3 | GDPD1 | 0.663567 | 4 | 0 | 3 |
| BCAS3 | BTG2 | 0.660373 | 3 | 0 | 3 |
| BCAS3 | DHX40 | 0.659321 | 6 | 0 | 6 |
For details and further investigation, click here