| Full name: blepharophimosis, epicanthus inversus and ptosis candidate 1 | Alias Symbol: NCRNA00187 | ||
| Type: non-coding RNA | Cytoband: 3q23 | ||
| Entrez ID: 60467 | HGNC ID: HGNC:13228 | Ensembl Gene: ENSG00000232416 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of BPESC1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BPESC1 | 60467 | 220772_at | -0.0361 | 0.8871 | |
| GSE20347 | BPESC1 | 60467 | 220772_at | 0.0194 | 0.6739 | |
| GSE23400 | BPESC1 | 60467 | 220772_at | -0.0177 | 0.2986 | |
| GSE26886 | BPESC1 | 60467 | 220772_at | -0.0203 | 0.8005 | |
| GSE29001 | BPESC1 | 60467 | 220772_at | 0.0081 | 0.9404 | |
| GSE38129 | BPESC1 | 60467 | 220772_at | 0.0194 | 0.5971 | |
| GSE45670 | BPESC1 | 60467 | 220772_at | 0.0802 | 0.3035 | |
| GSE53622 | BPESC1 | 60467 | 107548 | -0.6267 | 0.0005 | |
| GSE53624 | BPESC1 | 60467 | 107548 | -0.7307 | 0.0000 | |
| GSE63941 | BPESC1 | 60467 | 220772_at | -0.0744 | 0.5330 | |
| GSE77861 | BPESC1 | 60467 | 220772_at | 0.0115 | 0.9071 | |
| SRP133303 | BPESC1 | 60467 | RNAseq | -0.1632 | 0.6520 | |
| TCGA | BPESC1 | 60467 | RNAseq | 1.7542 | 0.3310 |
Upregulated datasets: 0; Downregulated datasets: 0.
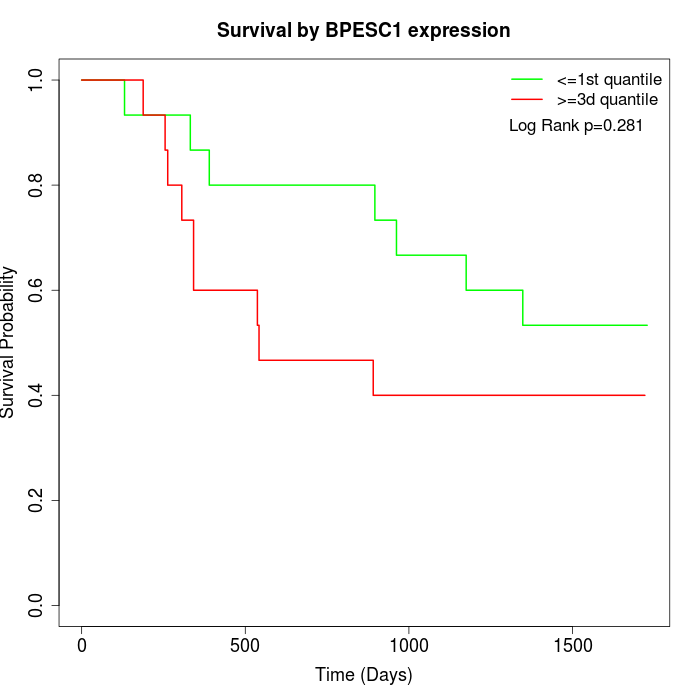
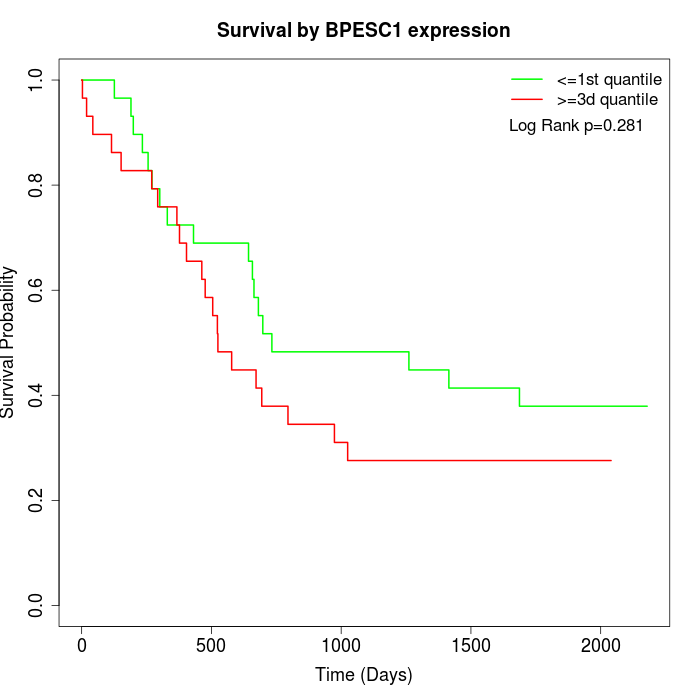
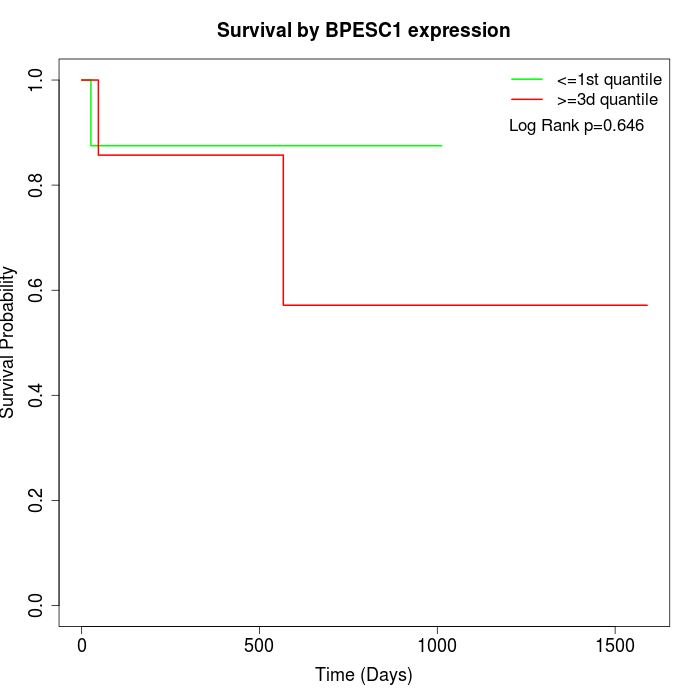
Survival by BPESC1 expression:
Note: Click image to view full size file.
Copy number change of BPESC1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BPESC1 | 60467 | 18 | 0 | 12 | |
| GSE20123 | BPESC1 | 60467 | 18 | 0 | 12 | |
| GSE43470 | BPESC1 | 60467 | 22 | 0 | 21 | |
| GSE46452 | BPESC1 | 60467 | 15 | 4 | 40 | |
| GSE47630 | BPESC1 | 60467 | 18 | 3 | 19 | |
| GSE54993 | BPESC1 | 60467 | 2 | 8 | 60 | |
| GSE54994 | BPESC1 | 60467 | 32 | 1 | 20 | |
| GSE60625 | BPESC1 | 60467 | 0 | 6 | 5 | |
| GSE74703 | BPESC1 | 60467 | 19 | 0 | 17 | |
| GSE74704 | BPESC1 | 60467 | 12 | 0 | 8 | |
| TCGA | BPESC1 | 60467 | 61 | 4 | 31 |
Total number of gains: 217; Total number of losses: 26; Total Number of normals: 245.
Somatic mutations of BPESC1:
Generating mutation plots.
Highly correlated genes for BPESC1:
Showing top 20/24 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BPESC1 | ANKRD26 | 0.694361 | 3 | 0 | 3 |
| BPESC1 | PDZD3 | 0.650957 | 3 | 0 | 3 |
| BPESC1 | KRTAP5-8 | 0.64288 | 3 | 0 | 3 |
| BPESC1 | KCNH6 | 0.635162 | 4 | 0 | 3 |
| BPESC1 | MAPRE3 | 0.633956 | 3 | 0 | 3 |
| BPESC1 | HDAC11 | 0.629471 | 3 | 0 | 3 |
| BPESC1 | RXRG | 0.629071 | 3 | 0 | 3 |
| BPESC1 | MLNR | 0.601985 | 3 | 0 | 3 |
| BPESC1 | BIRC7 | 0.599985 | 3 | 0 | 3 |
| BPESC1 | MEGF6 | 0.594313 | 4 | 0 | 3 |
| BPESC1 | HOXC5 | 0.587453 | 3 | 0 | 3 |
| BPESC1 | USP2 | 0.570117 | 3 | 0 | 3 |
| BPESC1 | AFP | 0.569041 | 4 | 0 | 3 |
| BPESC1 | IBA57 | 0.562181 | 3 | 0 | 3 |
| BPESC1 | NISCH | 0.551836 | 4 | 0 | 4 |
| BPESC1 | SMIM2 | 0.549871 | 5 | 0 | 3 |
| BPESC1 | KANSL3 | 0.547939 | 4 | 0 | 3 |
| BPESC1 | GRK4 | 0.539539 | 3 | 0 | 3 |
| BPESC1 | ADORA1 | 0.532613 | 4 | 0 | 3 |
| BPESC1 | SLC30A3 | 0.530348 | 4 | 0 | 3 |
For details and further investigation, click here