| Full name: BTG anti-proliferation factor 4 | Alias Symbol: PC3B|APRO3 | ||
| Type: protein-coding gene | Cytoband: 11q23.1 | ||
| Entrez ID: 54766 | HGNC ID: HGNC:13862 | Ensembl Gene: ENSG00000137707 | OMIM ID: 605673 |
| Drug and gene relationship at DGIdb | |||
Expression of BTG4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BTG4 | 54766 | 220766_at | 0.1660 | 0.3743 | |
| GSE20347 | BTG4 | 54766 | 220766_at | 0.0366 | 0.6367 | |
| GSE23400 | BTG4 | 54766 | 220766_at | -0.0359 | 0.3717 | |
| GSE26886 | BTG4 | 54766 | 220766_at | 0.2177 | 0.1701 | |
| GSE29001 | BTG4 | 54766 | 220766_at | -0.0227 | 0.8628 | |
| GSE38129 | BTG4 | 54766 | 220766_at | -0.0300 | 0.6708 | |
| GSE45670 | BTG4 | 54766 | 220766_at | 0.1275 | 0.3060 | |
| GSE53622 | BTG4 | 54766 | 77759 | 0.4795 | 0.0001 | |
| GSE53624 | BTG4 | 54766 | 77759 | 0.9102 | 0.0000 | |
| GSE63941 | BTG4 | 54766 | 220766_at | 0.1859 | 0.4566 | |
| GSE77861 | BTG4 | 54766 | 220766_at | 0.0859 | 0.3598 | |
| TCGA | BTG4 | 54766 | RNAseq | 3.1776 | 0.0068 |
Upregulated datasets: 1; Downregulated datasets: 0.
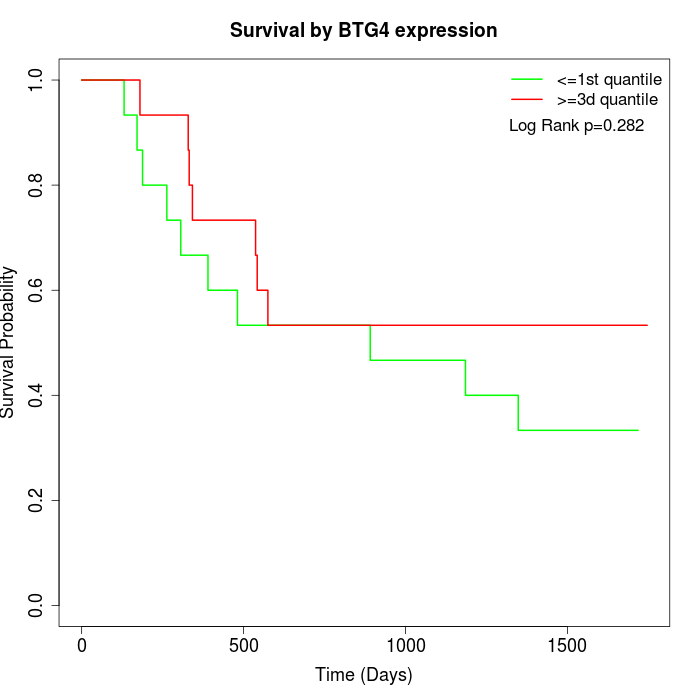
Survival by BTG4 expression:
Note: Click image to view full size file.
Copy number change of BTG4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BTG4 | 54766 | 0 | 12 | 18 | |
| GSE20123 | BTG4 | 54766 | 0 | 12 | 18 | |
| GSE43470 | BTG4 | 54766 | 2 | 6 | 35 | |
| GSE46452 | BTG4 | 54766 | 3 | 26 | 30 | |
| GSE47630 | BTG4 | 54766 | 2 | 19 | 19 | |
| GSE54993 | BTG4 | 54766 | 10 | 0 | 60 | |
| GSE54994 | BTG4 | 54766 | 5 | 19 | 29 | |
| GSE60625 | BTG4 | 54766 | 0 | 3 | 8 | |
| GSE74703 | BTG4 | 54766 | 1 | 4 | 31 | |
| GSE74704 | BTG4 | 54766 | 0 | 8 | 12 | |
| TCGA | BTG4 | 54766 | 6 | 49 | 41 |
Total number of gains: 29; Total number of losses: 158; Total Number of normals: 301.
Somatic mutations of BTG4:
Generating mutation plots.
Highly correlated genes for BTG4:
Showing top 20/541 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BTG4 | IFNA7 | 0.682595 | 6 | 0 | 6 |
| BTG4 | SMCP | 0.681448 | 5 | 0 | 5 |
| BTG4 | INCENP | 0.675793 | 3 | 0 | 3 |
| BTG4 | P2RY4 | 0.674405 | 6 | 0 | 6 |
| BTG4 | FRS3 | 0.659087 | 3 | 0 | 3 |
| BTG4 | TAS2R3 | 0.651657 | 3 | 0 | 3 |
| BTG4 | SPINK2 | 0.650394 | 4 | 0 | 4 |
| BTG4 | CGA | 0.649096 | 6 | 0 | 6 |
| BTG4 | IRS4 | 0.646795 | 3 | 0 | 3 |
| BTG4 | CLEC4M | 0.646196 | 5 | 0 | 4 |
| BTG4 | LGALS13 | 0.645478 | 4 | 0 | 3 |
| BTG4 | IFNA4 | 0.643933 | 4 | 0 | 4 |
| BTG4 | DNMT3L | 0.640138 | 4 | 0 | 4 |
| BTG4 | CCL1 | 0.637974 | 4 | 0 | 4 |
| BTG4 | KIR2DL2 | 0.633503 | 5 | 0 | 5 |
| BTG4 | CRH | 0.632705 | 3 | 0 | 3 |
| BTG4 | CA7 | 0.632145 | 5 | 0 | 4 |
| BTG4 | GK2 | 0.62952 | 4 | 0 | 3 |
| BTG4 | UTF1 | 0.628981 | 3 | 0 | 3 |
| BTG4 | AGTR2 | 0.628572 | 5 | 0 | 4 |
For details and further investigation, click here