| Full name: plexin C1 | Alias Symbol: VESPR|CD232 | ||
| Type: protein-coding gene | Cytoband: 12q22 | ||
| Entrez ID: 10154 | HGNC ID: HGNC:9106 | Ensembl Gene: ENSG00000136040 | OMIM ID: 604259 |
| Drug and gene relationship at DGIdb | |||
PLXNC1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance |
Expression of PLXNC1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PLXNC1 | 10154 | 213241_at | 0.5248 | 0.6891 | |
| GSE20347 | PLXNC1 | 10154 | 213241_at | 0.8621 | 0.0002 | |
| GSE23400 | PLXNC1 | 10154 | 213241_at | 0.7348 | 0.0000 | |
| GSE26886 | PLXNC1 | 10154 | 213241_at | 0.8473 | 0.0348 | |
| GSE29001 | PLXNC1 | 10154 | 213241_at | 0.7942 | 0.0622 | |
| GSE38129 | PLXNC1 | 10154 | 213241_at | 0.7788 | 0.0017 | |
| GSE45670 | PLXNC1 | 10154 | 213241_at | -0.1465 | 0.7698 | |
| GSE53622 | PLXNC1 | 10154 | 43686 | 1.1520 | 0.0000 | |
| GSE53624 | PLXNC1 | 10154 | 43686 | 1.1629 | 0.0000 | |
| GSE63941 | PLXNC1 | 10154 | 235328_at | 0.1213 | 0.7676 | |
| GSE77861 | PLXNC1 | 10154 | 235328_at | 0.2737 | 0.0548 | |
| GSE97050 | PLXNC1 | 10154 | A_23_P414958 | 0.7897 | 0.1227 | |
| SRP007169 | PLXNC1 | 10154 | RNAseq | 2.8459 | 0.0000 | |
| SRP008496 | PLXNC1 | 10154 | RNAseq | 2.0749 | 0.0000 | |
| SRP064894 | PLXNC1 | 10154 | RNAseq | 1.5224 | 0.0000 | |
| SRP133303 | PLXNC1 | 10154 | RNAseq | 1.1152 | 0.0000 | |
| SRP159526 | PLXNC1 | 10154 | RNAseq | 0.5609 | 0.3173 | |
| SRP193095 | PLXNC1 | 10154 | RNAseq | 0.6527 | 0.0000 | |
| SRP219564 | PLXNC1 | 10154 | RNAseq | 1.3773 | 0.0005 | |
| TCGA | PLXNC1 | 10154 | RNAseq | 0.2061 | 0.3316 |
Upregulated datasets: 7; Downregulated datasets: 0.
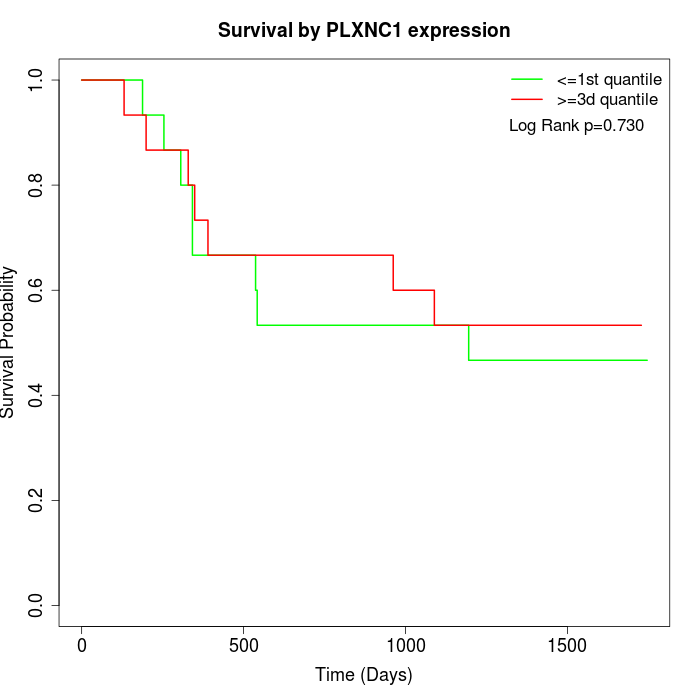
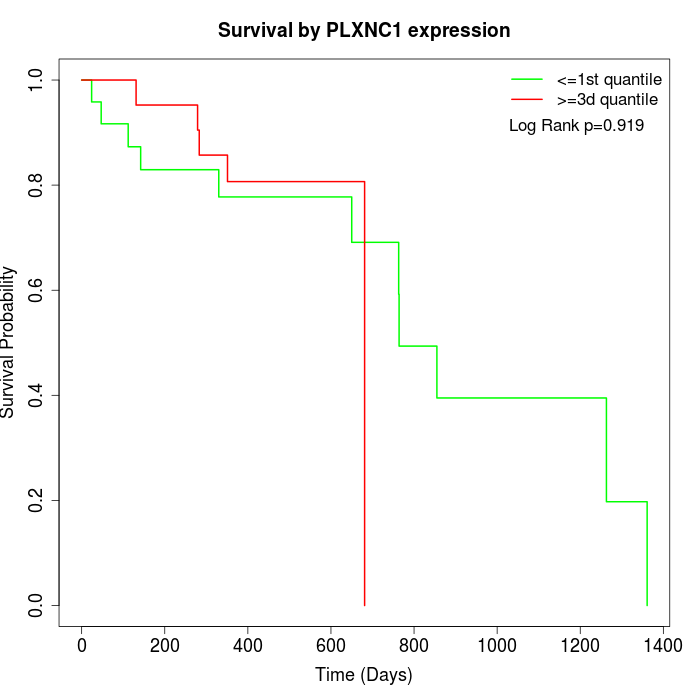
Survival by PLXNC1 expression:
Note: Click image to view full size file.
Copy number change of PLXNC1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PLXNC1 | 10154 | 4 | 2 | 24 | |
| GSE20123 | PLXNC1 | 10154 | 4 | 2 | 24 | |
| GSE43470 | PLXNC1 | 10154 | 3 | 0 | 40 | |
| GSE46452 | PLXNC1 | 10154 | 9 | 1 | 49 | |
| GSE47630 | PLXNC1 | 10154 | 10 | 1 | 29 | |
| GSE54993 | PLXNC1 | 10154 | 0 | 5 | 65 | |
| GSE54994 | PLXNC1 | 10154 | 4 | 1 | 48 | |
| GSE60625 | PLXNC1 | 10154 | 0 | 0 | 11 | |
| GSE74703 | PLXNC1 | 10154 | 3 | 0 | 33 | |
| GSE74704 | PLXNC1 | 10154 | 1 | 2 | 17 | |
| TCGA | PLXNC1 | 10154 | 17 | 10 | 69 |
Total number of gains: 55; Total number of losses: 24; Total Number of normals: 409.
Somatic mutations of PLXNC1:
Generating mutation plots.
Highly correlated genes for PLXNC1:
Showing top 20/1061 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PLXNC1 | GJD3 | 0.784651 | 3 | 0 | 3 |
| PLXNC1 | CMTM3 | 0.77848 | 6 | 0 | 6 |
| PLXNC1 | TYROBP | 0.771544 | 10 | 0 | 10 |
| PLXNC1 | PLEK | 0.75438 | 4 | 0 | 4 |
| PLXNC1 | IL2RA | 0.747024 | 4 | 0 | 4 |
| PLXNC1 | TMEM200A | 0.743565 | 6 | 0 | 6 |
| PLXNC1 | FPR3 | 0.740947 | 6 | 0 | 5 |
| PLXNC1 | RCN3 | 0.739834 | 6 | 0 | 6 |
| PLXNC1 | C1QC | 0.739032 | 5 | 0 | 5 |
| PLXNC1 | LAPTM5 | 0.736357 | 9 | 0 | 8 |
| PLXNC1 | PCOLCE | 0.734519 | 9 | 0 | 9 |
| PLXNC1 | FCGR3A | 0.734122 | 3 | 0 | 3 |
| PLXNC1 | C1QB | 0.7327 | 10 | 0 | 9 |
| PLXNC1 | FNDC1 | 0.726196 | 6 | 0 | 6 |
| PLXNC1 | PDIA5 | 0.724451 | 3 | 0 | 3 |
| PLXNC1 | NID2 | 0.723969 | 7 | 0 | 7 |
| PLXNC1 | OLFML2B | 0.72275 | 10 | 0 | 9 |
| PLXNC1 | CTSK | 0.720244 | 11 | 0 | 10 |
| PLXNC1 | CD300LF | 0.719427 | 4 | 0 | 4 |
| PLXNC1 | SULF1 | 0.718445 | 11 | 0 | 11 |
For details and further investigation, click here