| Full name: cancer antigen 1 | Alias Symbol: bA69L16.7|CT95 | ||
| Type: protein-coding gene | Cytoband: 6p24.3 | ||
| Entrez ID: 285782 | HGNC ID: HGNC:21622 | Ensembl Gene: ENSG00000164304 | OMIM ID: 608304 |
| Drug and gene relationship at DGIdb | |||
Expression of CAGE1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CAGE1 | 285782 | 1563787_a_at | 0.1520 | 0.4581 | |
| GSE26886 | CAGE1 | 285782 | 1563787_a_at | 0.2378 | 0.0061 | |
| GSE45670 | CAGE1 | 285782 | 1563787_a_at | 0.0576 | 0.2519 | |
| GSE53622 | CAGE1 | 285782 | 157589 | 2.6008 | 0.0000 | |
| GSE53624 | CAGE1 | 285782 | 157589 | 2.8654 | 0.0000 | |
| GSE63941 | CAGE1 | 285782 | 1563787_a_at | 0.1201 | 0.3309 | |
| GSE77861 | CAGE1 | 285782 | 1563787_a_at | -0.0116 | 0.9257 | |
| GSE97050 | CAGE1 | 285782 | A_33_P3326777 | 1.1410 | 0.1453 | |
| SRP133303 | CAGE1 | 285782 | RNAseq | 0.7245 | 0.0305 | |
| SRP159526 | CAGE1 | 285782 | RNAseq | 2.5132 | 0.0011 | |
| TCGA | CAGE1 | 285782 | RNAseq | 3.3224 | 0.0000 |
Upregulated datasets: 4; Downregulated datasets: 0.
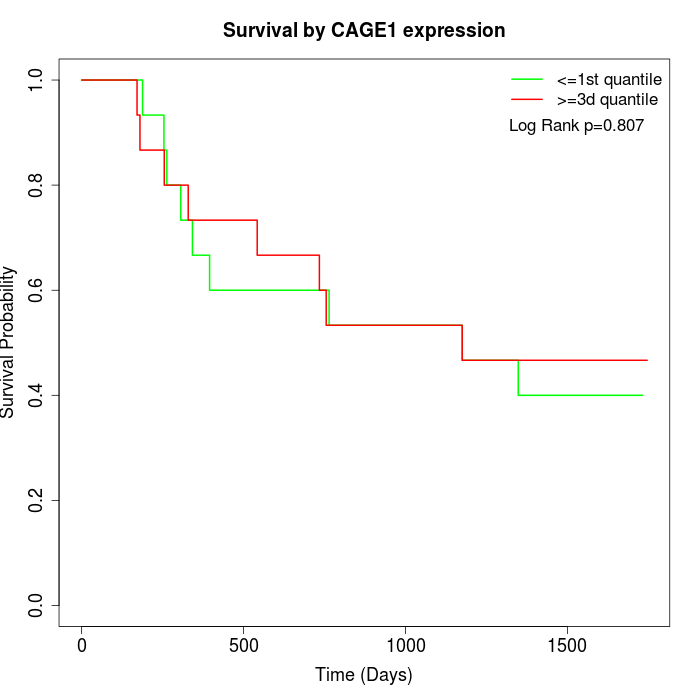
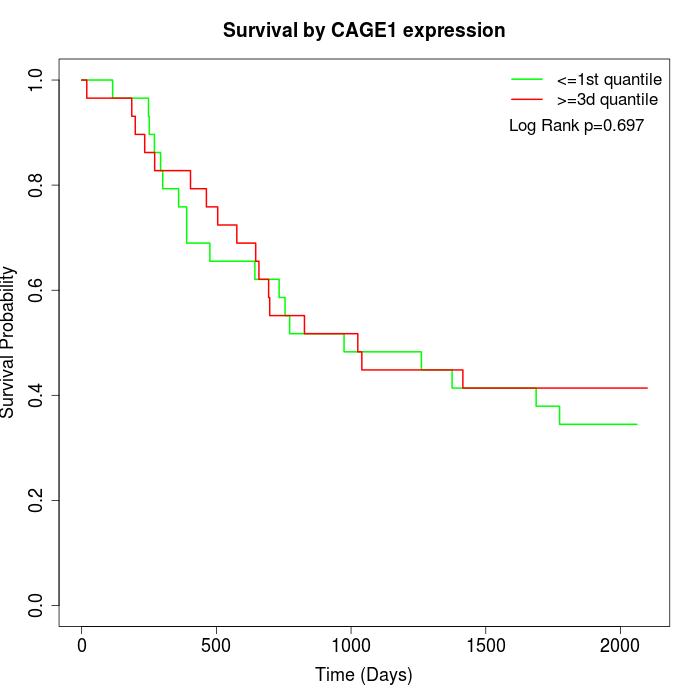
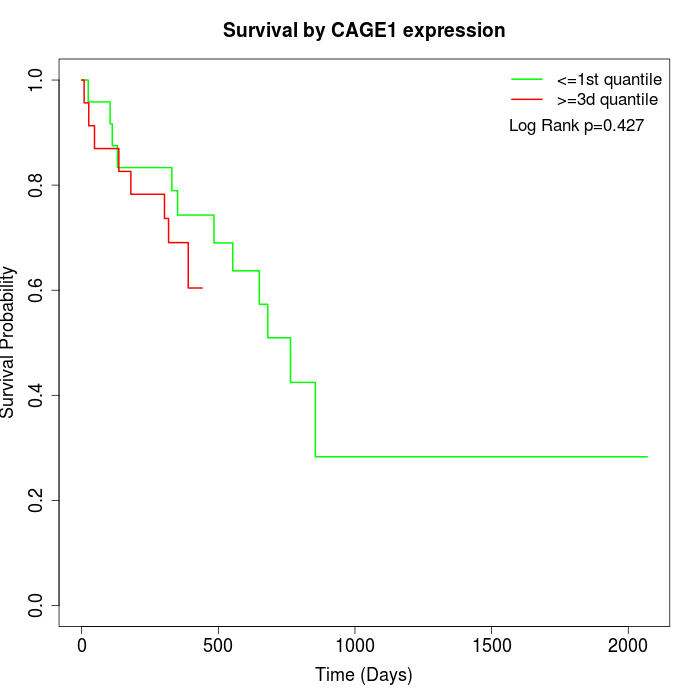
Survival by CAGE1 expression:
Note: Click image to view full size file.
Copy number change of CAGE1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CAGE1 | 285782 | 4 | 5 | 21 | |
| GSE20123 | CAGE1 | 285782 | 4 | 5 | 21 | |
| GSE43470 | CAGE1 | 285782 | 10 | 2 | 31 | |
| GSE46452 | CAGE1 | 285782 | 1 | 10 | 48 | |
| GSE47630 | CAGE1 | 285782 | 7 | 8 | 25 | |
| GSE54993 | CAGE1 | 285782 | 1 | 1 | 68 | |
| GSE54994 | CAGE1 | 285782 | 10 | 3 | 40 | |
| GSE60625 | CAGE1 | 285782 | 0 | 1 | 10 | |
| GSE74703 | CAGE1 | 285782 | 8 | 2 | 26 | |
| GSE74704 | CAGE1 | 285782 | 2 | 2 | 16 | |
| TCGA | CAGE1 | 285782 | 15 | 29 | 52 |
Total number of gains: 62; Total number of losses: 68; Total Number of normals: 358.
Somatic mutations of CAGE1:
Generating mutation plots.
Highly correlated genes for CAGE1:
Showing top 20/729 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CAGE1 | MND1 | 0.719616 | 3 | 0 | 3 |
| CAGE1 | RBL1 | 0.717588 | 4 | 0 | 4 |
| CAGE1 | DKC1 | 0.717499 | 3 | 0 | 3 |
| CAGE1 | FOXRED2 | 0.714664 | 3 | 0 | 3 |
| CAGE1 | DHFR | 0.710589 | 4 | 0 | 4 |
| CAGE1 | MAD2L2 | 0.706029 | 3 | 0 | 3 |
| CAGE1 | ANLN | 0.702119 | 4 | 0 | 4 |
| CAGE1 | KIF14 | 0.699454 | 4 | 0 | 4 |
| CAGE1 | CHST1 | 0.692702 | 5 | 0 | 5 |
| CAGE1 | NDE1 | 0.692614 | 3 | 0 | 3 |
| CAGE1 | PCGF1 | 0.686204 | 4 | 0 | 4 |
| CAGE1 | FAM89A | 0.682996 | 3 | 0 | 3 |
| CAGE1 | HOXD11 | 0.68236 | 5 | 0 | 5 |
| CAGE1 | NOP56 | 0.681769 | 3 | 0 | 3 |
| CAGE1 | CHEK1 | 0.676229 | 4 | 0 | 4 |
| CAGE1 | DLL3 | 0.67555 | 4 | 0 | 3 |
| CAGE1 | SLC25A17 | 0.6755 | 4 | 0 | 4 |
| CAGE1 | TREM2 | 0.675098 | 5 | 0 | 4 |
| CAGE1 | PYCR2 | 0.674918 | 3 | 0 | 3 |
| CAGE1 | ZNF484 | 0.673219 | 3 | 0 | 3 |
For details and further investigation, click here