| Full name: nudE neurodevelopment protein 1 | Alias Symbol: nudE|FLJ20101|NDE | ||
| Type: protein-coding gene | Cytoband: 16p13.11 | ||
| Entrez ID: 54820 | HGNC ID: HGNC:17619 | Ensembl Gene: ENSG00000072864 | OMIM ID: 609449 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of NDE1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NDE1 | 54820 | 227249_at | 0.7977 | 0.1793 | |
| GSE20347 | NDE1 | 54820 | 218414_s_at | 0.8771 | 0.0000 | |
| GSE23400 | NDE1 | 54820 | 218414_s_at | 0.5216 | 0.0000 | |
| GSE26886 | NDE1 | 54820 | 227249_at | 1.0389 | 0.0088 | |
| GSE29001 | NDE1 | 54820 | 218414_s_at | 0.8452 | 0.0010 | |
| GSE38129 | NDE1 | 54820 | 218414_s_at | 0.6875 | 0.0001 | |
| GSE45670 | NDE1 | 54820 | 227249_at | 1.1015 | 0.0002 | |
| GSE53622 | NDE1 | 54820 | 3016 | 0.8169 | 0.0000 | |
| GSE53624 | NDE1 | 54820 | 3016 | 0.9825 | 0.0000 | |
| GSE63941 | NDE1 | 54820 | 227249_at | 1.3278 | 0.0259 | |
| GSE77861 | NDE1 | 54820 | 218414_s_at | 1.2260 | 0.0034 | |
| GSE97050 | NDE1 | 54820 | A_23_P206901 | 0.8625 | 0.1186 | |
| SRP007169 | NDE1 | 54820 | RNAseq | 1.5662 | 0.0001 | |
| SRP008496 | NDE1 | 54820 | RNAseq | 1.0857 | 0.0005 | |
| SRP064894 | NDE1 | 54820 | RNAseq | 0.6193 | 0.0024 | |
| SRP133303 | NDE1 | 54820 | RNAseq | 0.8323 | 0.0000 | |
| SRP159526 | NDE1 | 54820 | RNAseq | 0.5981 | 0.0166 | |
| SRP193095 | NDE1 | 54820 | RNAseq | 1.1965 | 0.0000 | |
| SRP219564 | NDE1 | 54820 | RNAseq | 0.7767 | 0.0375 | |
| TCGA | NDE1 | 54820 | RNAseq | 0.1193 | 0.0782 |
Upregulated datasets: 7; Downregulated datasets: 0.
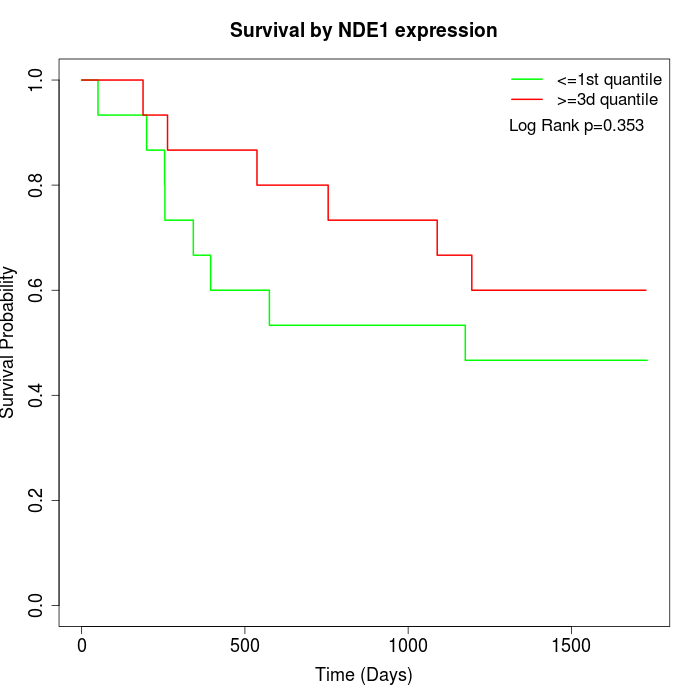
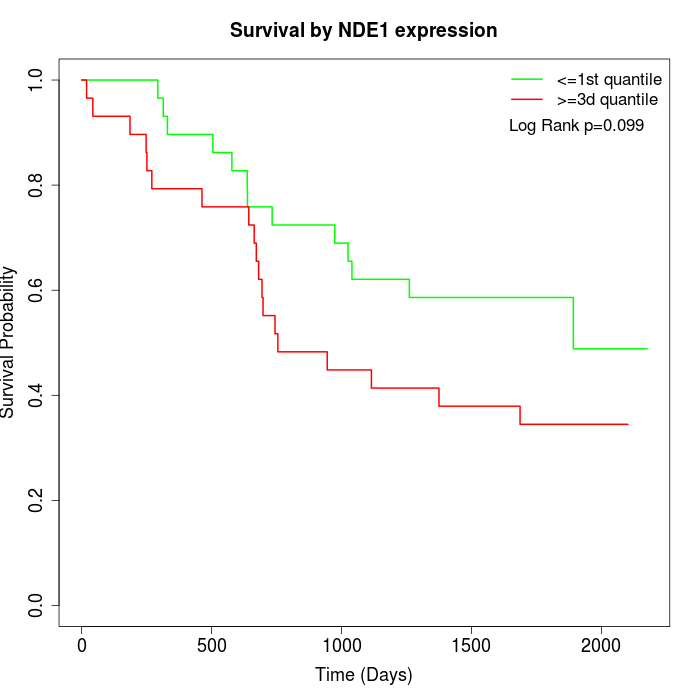
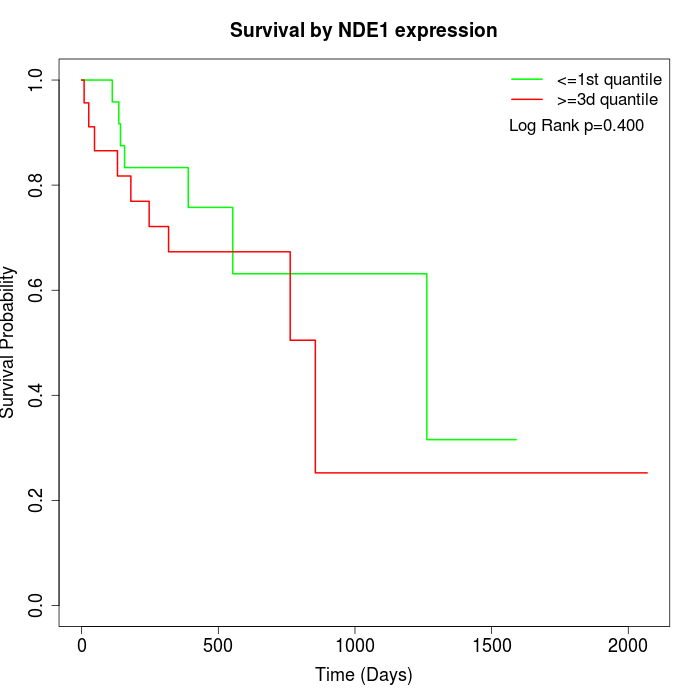
Survival by NDE1 expression:
Note: Click image to view full size file.
Copy number change of NDE1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NDE1 | 54820 | 5 | 5 | 20 | |
| GSE20123 | NDE1 | 54820 | 5 | 4 | 21 | |
| GSE43470 | NDE1 | 54820 | 3 | 3 | 37 | |
| GSE46452 | NDE1 | 54820 | 38 | 1 | 20 | |
| GSE47630 | NDE1 | 54820 | 11 | 6 | 23 | |
| GSE54993 | NDE1 | 54820 | 3 | 5 | 62 | |
| GSE54994 | NDE1 | 54820 | 4 | 10 | 39 | |
| GSE60625 | NDE1 | 54820 | 4 | 1 | 6 | |
| GSE74703 | NDE1 | 54820 | 3 | 2 | 31 | |
| GSE74704 | NDE1 | 54820 | 2 | 1 | 17 | |
| TCGA | NDE1 | 54820 | 20 | 12 | 64 |
Total number of gains: 98; Total number of losses: 50; Total Number of normals: 340.
Somatic mutations of NDE1:
Generating mutation plots.
Highly correlated genes for NDE1:
Showing top 20/1561 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NDE1 | CDCA2 | 0.740962 | 6 | 0 | 6 |
| NDE1 | RNPS1 | 0.740519 | 11 | 0 | 11 |
| NDE1 | PKMYT1 | 0.733273 | 9 | 0 | 9 |
| NDE1 | EBNA1BP2 | 0.733213 | 3 | 0 | 3 |
| NDE1 | TTL | 0.721411 | 6 | 0 | 5 |
| NDE1 | ALKBH6 | 0.721333 | 4 | 0 | 4 |
| NDE1 | PDE7A | 0.720894 | 6 | 0 | 6 |
| NDE1 | TMEM132A | 0.718567 | 8 | 0 | 8 |
| NDE1 | YWHAG | 0.711465 | 6 | 0 | 6 |
| NDE1 | SPNS1 | 0.708227 | 5 | 0 | 5 |
| NDE1 | SPON2 | 0.703546 | 3 | 0 | 3 |
| NDE1 | SNUPN | 0.703344 | 3 | 0 | 3 |
| NDE1 | AURKA | 0.703312 | 12 | 0 | 11 |
| NDE1 | HHEX | 0.700174 | 3 | 0 | 3 |
| NDE1 | UBE2C | 0.697594 | 12 | 0 | 11 |
| NDE1 | GTSE1 | 0.697327 | 11 | 0 | 10 |
| NDE1 | NOP56 | 0.694392 | 3 | 0 | 3 |
| NDE1 | EME1 | 0.693286 | 6 | 0 | 6 |
| NDE1 | CAGE1 | 0.692614 | 3 | 0 | 3 |
| NDE1 | SLC7A6 | 0.691247 | 8 | 0 | 8 |
For details and further investigation, click here