| Full name: checkpoint kinase 1 | Alias Symbol: CHK1 | ||
| Type: protein-coding gene | Cytoband: 11q24.2 | ||
| Entrez ID: 1111 | HGNC ID: HGNC:1925 | Ensembl Gene: ENSG00000149554 | OMIM ID: 603078 |
| Drug and gene relationship at DGIdb | |||
CHEK1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle | |
| hsa04115 | p53 signaling pathway | |
| hsa05166 | HTLV-I infection |
Expression of CHEK1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CHEK1 | 1111 | 205394_at | 1.3877 | 0.0286 | |
| GSE20347 | CHEK1 | 1111 | 205394_at | 1.2126 | 0.0000 | |
| GSE23400 | CHEK1 | 1111 | 205394_at | 0.6936 | 0.0000 | |
| GSE26886 | CHEK1 | 1111 | 205394_at | 0.8404 | 0.0205 | |
| GSE29001 | CHEK1 | 1111 | 205394_at | 1.5644 | 0.0013 | |
| GSE38129 | CHEK1 | 1111 | 205394_at | 1.4181 | 0.0000 | |
| GSE45670 | CHEK1 | 1111 | 238075_at | 1.2451 | 0.0000 | |
| GSE53622 | CHEK1 | 1111 | 38020 | 1.3384 | 0.0000 | |
| GSE53624 | CHEK1 | 1111 | 38020 | 1.5845 | 0.0000 | |
| GSE63941 | CHEK1 | 1111 | 205394_at | 1.6738 | 0.0028 | |
| GSE77861 | CHEK1 | 1111 | 205393_s_at | 0.6880 | 0.0950 | |
| GSE97050 | CHEK1 | 1111 | A_33_P3349536 | 1.4374 | 0.1555 | |
| SRP007169 | CHEK1 | 1111 | RNAseq | 1.5126 | 0.0009 | |
| SRP008496 | CHEK1 | 1111 | RNAseq | 1.7770 | 0.0000 | |
| SRP064894 | CHEK1 | 1111 | RNAseq | 1.0213 | 0.0000 | |
| SRP133303 | CHEK1 | 1111 | RNAseq | 0.9699 | 0.0000 | |
| SRP159526 | CHEK1 | 1111 | RNAseq | 0.7945 | 0.0112 | |
| SRP193095 | CHEK1 | 1111 | RNAseq | 0.5205 | 0.0003 | |
| SRP219564 | CHEK1 | 1111 | RNAseq | 0.8456 | 0.0458 | |
| TCGA | CHEK1 | 1111 | RNAseq | 0.8594 | 0.0000 |
Upregulated datasets: 11; Downregulated datasets: 0.
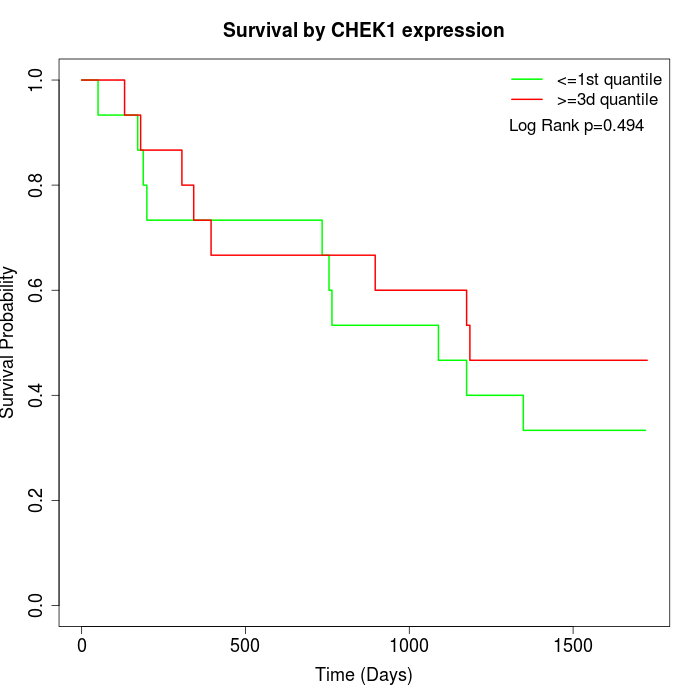
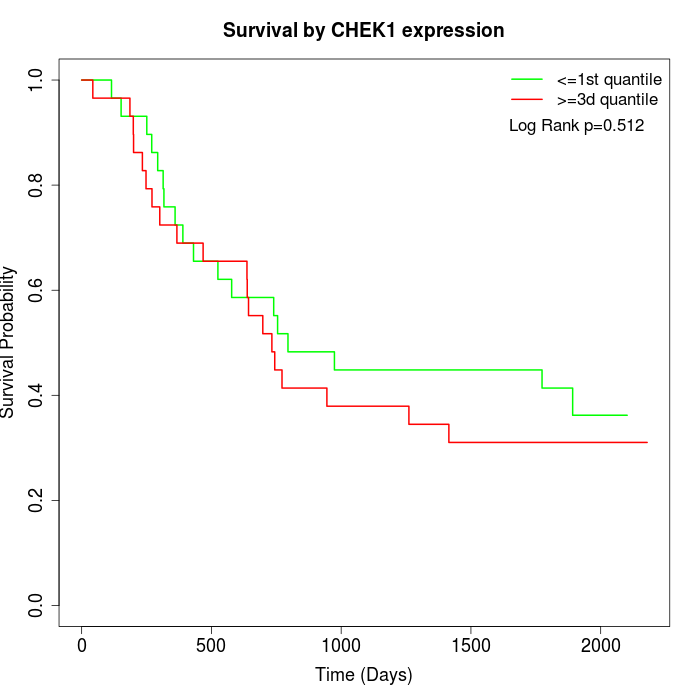
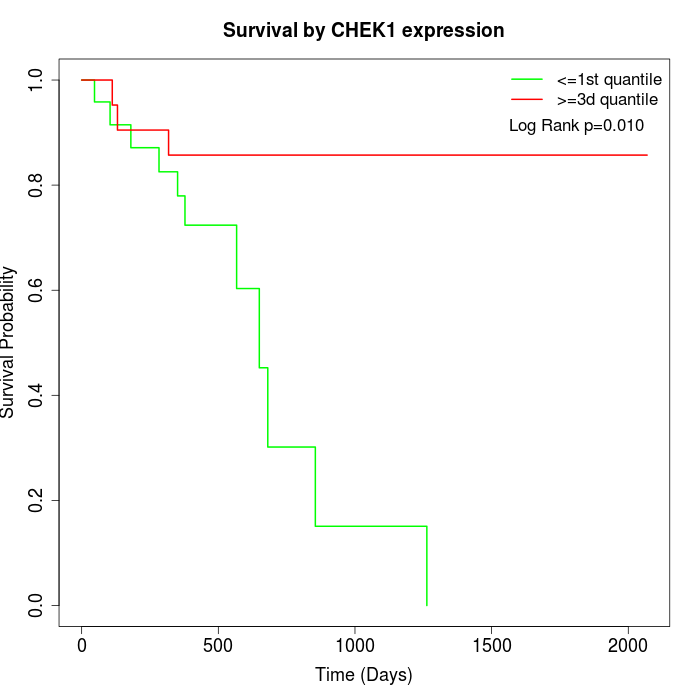
Survival by CHEK1 expression:
Note: Click image to view full size file.
Copy number change of CHEK1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CHEK1 | 1111 | 0 | 13 | 17 | |
| GSE20123 | CHEK1 | 1111 | 0 | 13 | 17 | |
| GSE43470 | CHEK1 | 1111 | 2 | 7 | 34 | |
| GSE46452 | CHEK1 | 1111 | 3 | 26 | 30 | |
| GSE47630 | CHEK1 | 1111 | 2 | 19 | 19 | |
| GSE54993 | CHEK1 | 1111 | 10 | 0 | 60 | |
| GSE54994 | CHEK1 | 1111 | 5 | 19 | 29 | |
| GSE60625 | CHEK1 | 1111 | 0 | 3 | 8 | |
| GSE74703 | CHEK1 | 1111 | 1 | 5 | 30 | |
| GSE74704 | CHEK1 | 1111 | 0 | 9 | 11 | |
| TCGA | CHEK1 | 1111 | 7 | 50 | 39 |
Total number of gains: 30; Total number of losses: 164; Total Number of normals: 294.
Somatic mutations of CHEK1:
Generating mutation plots.
Highly correlated genes for CHEK1:
Showing top 20/1691 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CHEK1 | CDK1 | 0.804907 | 12 | 0 | 12 |
| CHEK1 | DDIAS | 0.799437 | 7 | 0 | 7 |
| CHEK1 | TTK | 0.799357 | 12 | 0 | 12 |
| CHEK1 | CDCA5 | 0.798104 | 7 | 0 | 7 |
| CHEK1 | RAD51AP1 | 0.796451 | 13 | 0 | 13 |
| CHEK1 | GMNN | 0.795058 | 13 | 0 | 13 |
| CHEK1 | DLGAP5 | 0.794669 | 13 | 0 | 13 |
| CHEK1 | KIF23 | 0.794602 | 13 | 0 | 13 |
| CHEK1 | FEN1 | 0.79439 | 9 | 0 | 9 |
| CHEK1 | FOXM1 | 0.794016 | 12 | 0 | 12 |
| CHEK1 | CDC20 | 0.793904 | 11 | 0 | 10 |
| CHEK1 | CDKN3 | 0.789504 | 13 | 0 | 13 |
| CHEK1 | ATAD2 | 0.789085 | 13 | 0 | 13 |
| CHEK1 | CDCA2 | 0.786618 | 7 | 0 | 7 |
| CHEK1 | BUB1 | 0.78565 | 11 | 0 | 11 |
| CHEK1 | MELK | 0.785581 | 13 | 0 | 13 |
| CHEK1 | KIF2C | 0.783665 | 12 | 0 | 12 |
| CHEK1 | HMMR | 0.783612 | 12 | 0 | 11 |
| CHEK1 | TPX2 | 0.782898 | 13 | 0 | 11 |
| CHEK1 | CEP55 | 0.782829 | 13 | 0 | 13 |
For details and further investigation, click here