| Full name: caspase 1 | Alias Symbol: ICE | ||
| Type: protein-coding gene | Cytoband: 11q22.3 | ||
| Entrez ID: 834 | HGNC ID: HGNC:1499 | Ensembl Gene: ENSG00000137752 | OMIM ID: 147678 |
| Drug and gene relationship at DGIdb | |||
CASP1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04621 | NOD-like receptor signaling pathway | |
| hsa05132 | Salmonella infection | |
| hsa05133 | Pertussis | |
| hsa05134 | Legionellosis | |
| hsa05164 | Influenza A |
Expression of CASP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CASP1 | 834 | 209970_x_at | -0.1865 | 0.8046 | |
| GSE20347 | CASP1 | 834 | 209970_x_at | -0.6353 | 0.0228 | |
| GSE23400 | CASP1 | 834 | 211366_x_at | 0.0911 | 0.4421 | |
| GSE26886 | CASP1 | 834 | 209970_x_at | -0.7549 | 0.0157 | |
| GSE29001 | CASP1 | 834 | 209970_x_at | -0.1405 | 0.7817 | |
| GSE38129 | CASP1 | 834 | 209970_x_at | -0.2296 | 0.3537 | |
| GSE45670 | CASP1 | 834 | 209970_x_at | 0.5406 | 0.0186 | |
| GSE53622 | CASP1 | 834 | 15327 | 0.0679 | 0.5586 | |
| GSE53624 | CASP1 | 834 | 15327 | -0.1063 | 0.3617 | |
| GSE63941 | CASP1 | 834 | 209970_x_at | -0.8364 | 0.5276 | |
| GSE77861 | CASP1 | 834 | 209970_x_at | -0.8150 | 0.0989 | |
| GSE97050 | CASP1 | 834 | A_23_P202978 | 0.5343 | 0.3596 | |
| SRP007169 | CASP1 | 834 | RNAseq | -1.0897 | 0.0032 | |
| SRP008496 | CASP1 | 834 | RNAseq | -1.0230 | 0.0000 | |
| SRP064894 | CASP1 | 834 | RNAseq | 0.0205 | 0.9159 | |
| SRP133303 | CASP1 | 834 | RNAseq | -0.0764 | 0.7765 | |
| SRP159526 | CASP1 | 834 | RNAseq | -0.4206 | 0.4374 | |
| SRP193095 | CASP1 | 834 | RNAseq | -0.5725 | 0.0000 | |
| SRP219564 | CASP1 | 834 | RNAseq | 0.4677 | 0.3681 | |
| TCGA | CASP1 | 834 | RNAseq | 0.5843 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 2.
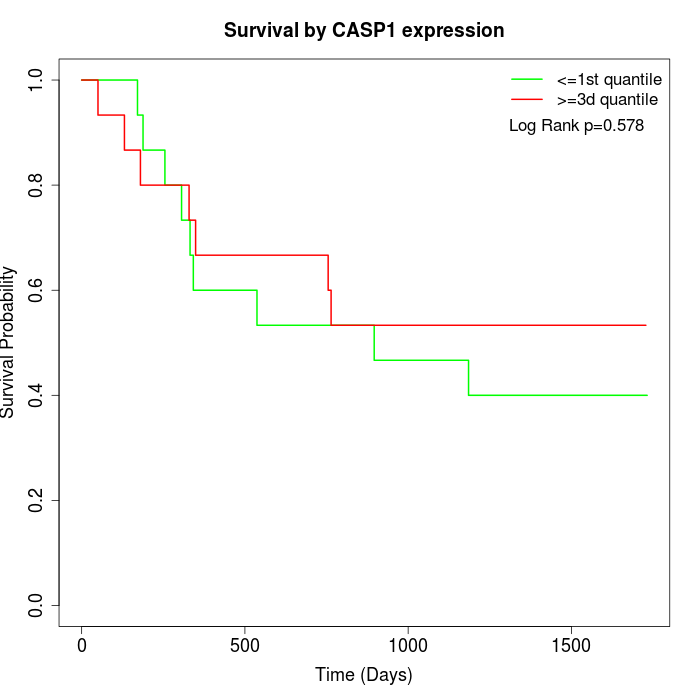
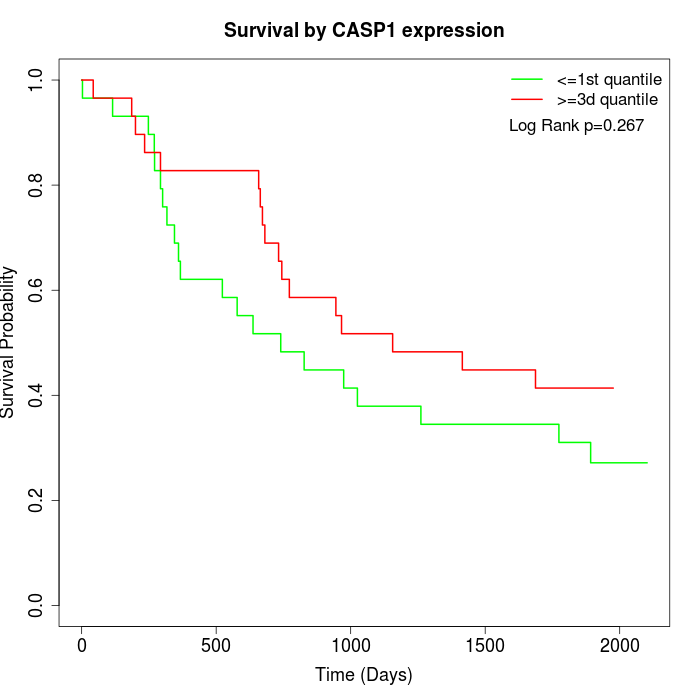
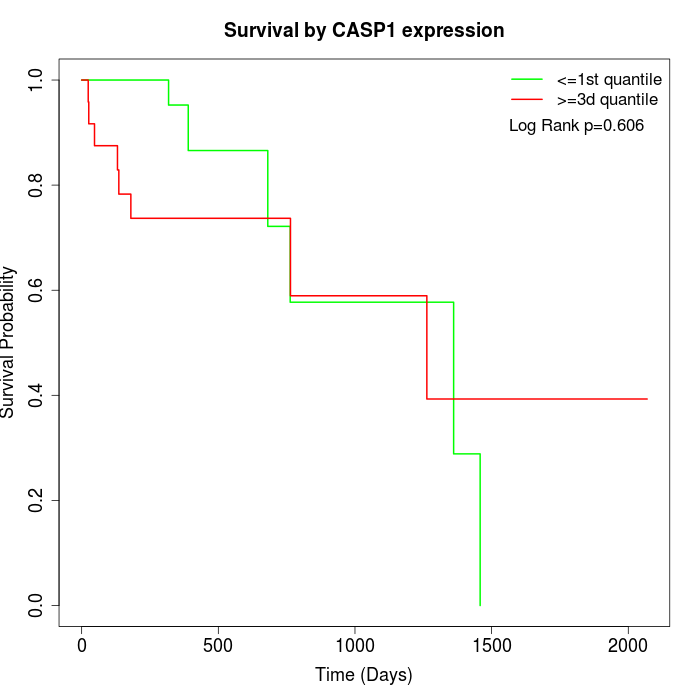
Survival by CASP1 expression:
Note: Click image to view full size file.
Copy number change of CASP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CASP1 | 834 | 1 | 11 | 18 | |
| GSE20123 | CASP1 | 834 | 1 | 11 | 18 | |
| GSE43470 | CASP1 | 834 | 3 | 6 | 34 | |
| GSE46452 | CASP1 | 834 | 3 | 26 | 30 | |
| GSE47630 | CASP1 | 834 | 2 | 19 | 19 | |
| GSE54993 | CASP1 | 834 | 9 | 1 | 60 | |
| GSE54994 | CASP1 | 834 | 4 | 19 | 30 | |
| GSE60625 | CASP1 | 834 | 0 | 3 | 8 | |
| GSE74703 | CASP1 | 834 | 2 | 4 | 30 | |
| GSE74704 | CASP1 | 834 | 1 | 7 | 12 | |
| TCGA | CASP1 | 834 | 9 | 44 | 43 |
Total number of gains: 35; Total number of losses: 151; Total Number of normals: 302.
Somatic mutations of CASP1:
Generating mutation plots.
Highly correlated genes for CASP1:
Showing top 20/599 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CASP1 | CASP4 | 0.76564 | 13 | 0 | 12 |
| CASP1 | CARD17 | 0.757745 | 4 | 0 | 3 |
| CASP1 | HSBP1L1 | 0.741542 | 3 | 0 | 3 |
| CASP1 | GFM2 | 0.724764 | 3 | 0 | 3 |
| CASP1 | PRKAR2A | 0.721601 | 3 | 0 | 3 |
| CASP1 | TMEM126A | 0.71393 | 5 | 0 | 4 |
| CASP1 | NAA20 | 0.705781 | 4 | 0 | 4 |
| CASP1 | CERS3 | 0.705621 | 5 | 0 | 4 |
| CASP1 | SLC35A4 | 0.704646 | 3 | 0 | 3 |
| CASP1 | HDHD2 | 0.698056 | 3 | 0 | 3 |
| CASP1 | IPMK | 0.697407 | 3 | 0 | 3 |
| CASP1 | CNFN | 0.6933 | 3 | 0 | 3 |
| CASP1 | CARD16 | 0.686415 | 9 | 0 | 7 |
| CASP1 | QSOX1 | 0.681981 | 3 | 0 | 3 |
| CASP1 | MRPL48 | 0.680846 | 3 | 0 | 3 |
| CASP1 | ZNF823 | 0.679177 | 3 | 0 | 3 |
| CASP1 | OXNAD1 | 0.678897 | 3 | 0 | 3 |
| CASP1 | SLC38A9 | 0.678837 | 4 | 0 | 4 |
| CASP1 | PPP4R2 | 0.677558 | 3 | 0 | 3 |
| CASP1 | CYP4X1 | 0.675451 | 4 | 0 | 4 |
For details and further investigation, click here