| Full name: C-C motif chemokine ligand 3 | Alias Symbol: G0S19-1|LD78ALPHA|MIP-1-alpha | ||
| Type: protein-coding gene | Cytoband: 17q12 | ||
| Entrez ID: 6348 | HGNC ID: HGNC:10627 | Ensembl Gene: ENSG00000277632 | OMIM ID: 182283 |
| Drug and gene relationship at DGIdb | |||
CCL3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway | |
| hsa04620 | Toll-like receptor signaling pathway |
Expression of CCL3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | CCL3 | 6348 | 13320 | -0.0848 | 0.6524 | |
| GSE53624 | CCL3 | 6348 | 13320 | -0.2114 | 0.1924 | |
| GSE97050 | CCL3 | 6348 | A_33_P3316273 | 0.4806 | 0.2470 | |
| SRP064894 | CCL3 | 6348 | RNAseq | 2.8267 | 0.0000 | |
| SRP133303 | CCL3 | 6348 | RNAseq | 2.8574 | 0.0000 | |
| SRP159526 | CCL3 | 6348 | RNAseq | 2.1462 | 0.0002 | |
| SRP193095 | CCL3 | 6348 | RNAseq | 3.3999 | 0.0000 | |
| SRP219564 | CCL3 | 6348 | RNAseq | 1.9867 | 0.0504 | |
| TCGA | CCL3 | 6348 | RNAseq | 1.9737 | 0.0000 |
Upregulated datasets: 5; Downregulated datasets: 0.
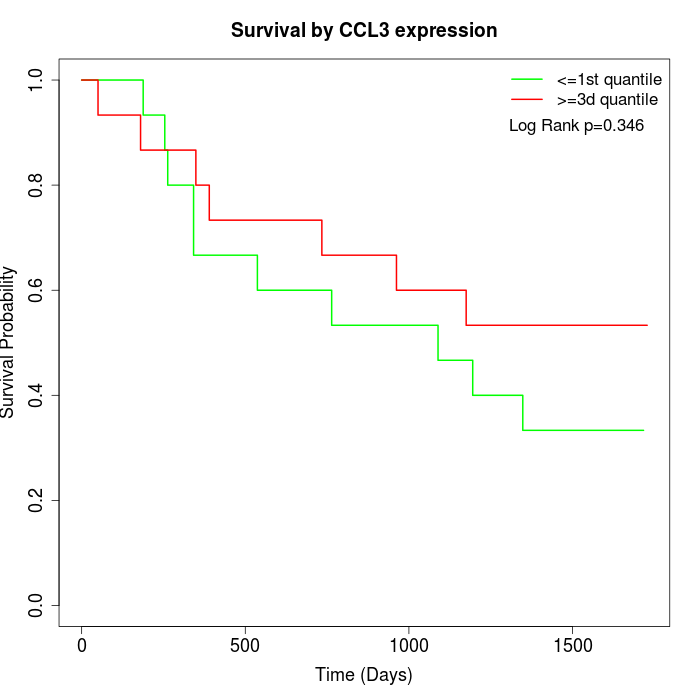
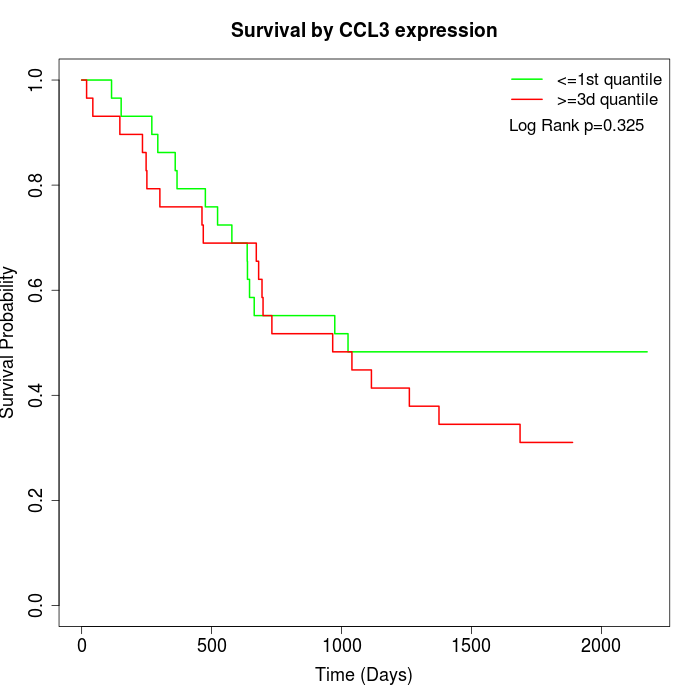
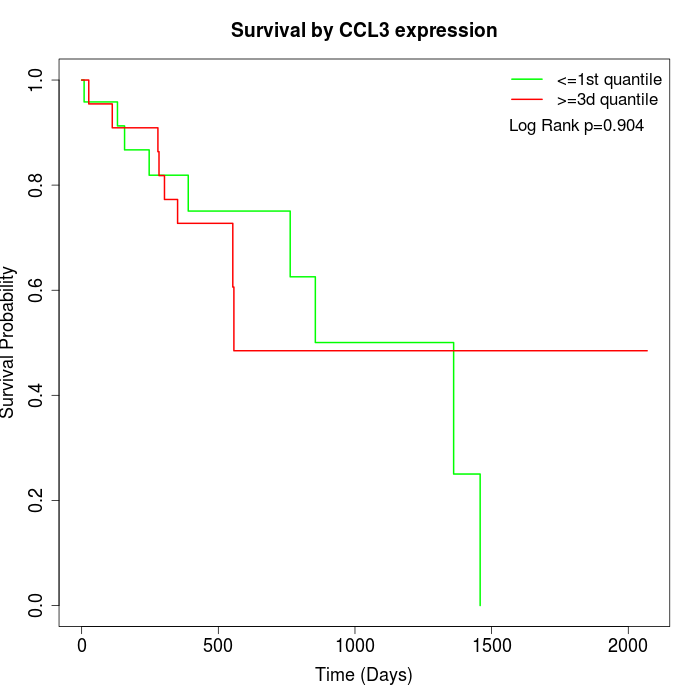
Survival by CCL3 expression:
Note: Click image to view full size file.
Copy number change of CCL3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCL3 | 6348 | 5 | 1 | 24 | |
| GSE20123 | CCL3 | 6348 | 5 | 1 | 24 | |
| GSE43470 | CCL3 | 6348 | 1 | 2 | 40 | |
| GSE46452 | CCL3 | 6348 | 34 | 0 | 25 | |
| GSE47630 | CCL3 | 6348 | 7 | 1 | 32 | |
| GSE54993 | CCL3 | 6348 | 3 | 3 | 64 | |
| GSE54994 | CCL3 | 6348 | 8 | 6 | 39 | |
| GSE60625 | CCL3 | 6348 | 4 | 0 | 7 | |
| GSE74703 | CCL3 | 6348 | 1 | 1 | 34 | |
| GSE74704 | CCL3 | 6348 | 3 | 1 | 16 | |
| TCGA | CCL3 | 6348 | 20 | 9 | 67 |
Total number of gains: 91; Total number of losses: 25; Total Number of normals: 372.
Somatic mutations of CCL3:
Generating mutation plots.
Highly correlated genes for CCL3:
Showing top 20/136 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCL3 | MNDA | 0.770077 | 3 | 0 | 3 |
| CCL3 | MRC1 | 0.754596 | 3 | 0 | 3 |
| CCL3 | LRRC25 | 0.743575 | 3 | 0 | 3 |
| CCL3 | FCGR2B | 0.726281 | 3 | 0 | 3 |
| CCL3 | DPEP2 | 0.720609 | 3 | 0 | 3 |
| CCL3 | CMKLR1 | 0.717664 | 3 | 0 | 3 |
| CCL3 | LILRB5 | 0.713259 | 3 | 0 | 3 |
| CCL3 | MPEG1 | 0.709108 | 3 | 0 | 3 |
| CCL3 | CSF2RB | 0.708609 | 3 | 0 | 3 |
| CCL3 | LCP2 | 0.707739 | 3 | 0 | 3 |
| CCL3 | C1orf162 | 0.697496 | 3 | 0 | 3 |
| CCL3 | LILRB2 | 0.681831 | 3 | 0 | 3 |
| CCL3 | C19orf38 | 0.681227 | 3 | 0 | 3 |
| CCL3 | CCL8 | 0.679317 | 4 | 0 | 4 |
| CCL3 | STAB1 | 0.677381 | 3 | 0 | 3 |
| CCL3 | RAB8B | 0.673732 | 3 | 0 | 3 |
| CCL3 | PDCD1LG2 | 0.665271 | 3 | 0 | 3 |
| CCL3 | ATP8B4 | 0.661377 | 3 | 0 | 3 |
| CCL3 | EVI2B | 0.660208 | 3 | 0 | 3 |
| CCL3 | SIGLEC1 | 0.660107 | 3 | 0 | 3 |
For details and further investigation, click here