| Full name: copper chaperone for superoxide dismutase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11q13.2 | ||
| Entrez ID: 9973 | HGNC ID: HGNC:1613 | Ensembl Gene: ENSG00000173992 | OMIM ID: 603864 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CCS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCS | 9973 | 203522_at | 0.1925 | 0.3293 | |
| GSE20347 | CCS | 9973 | 203522_at | 0.3703 | 0.0066 | |
| GSE23400 | CCS | 9973 | 203522_at | 0.1287 | 0.0659 | |
| GSE26886 | CCS | 9973 | 203522_at | 0.4204 | 0.0716 | |
| GSE29001 | CCS | 9973 | 203522_at | 0.2872 | 0.3957 | |
| GSE38129 | CCS | 9973 | 203522_at | 0.1957 | 0.2357 | |
| GSE45670 | CCS | 9973 | 203522_at | 0.1094 | 0.4625 | |
| GSE53622 | CCS | 9973 | 74586 | 0.0517 | 0.5171 | |
| GSE53624 | CCS | 9973 | 74586 | 0.1202 | 0.0719 | |
| GSE63941 | CCS | 9973 | 203522_at | 0.1826 | 0.7020 | |
| GSE77861 | CCS | 9973 | 203522_at | 0.1699 | 0.4225 | |
| GSE97050 | CCS | 9973 | A_23_P127475 | -0.0488 | 0.8732 | |
| SRP007169 | CCS | 9973 | RNAseq | 0.2858 | 0.6085 | |
| SRP008496 | CCS | 9973 | RNAseq | -0.0609 | 0.8710 | |
| SRP064894 | CCS | 9973 | RNAseq | 0.3488 | 0.1369 | |
| SRP133303 | CCS | 9973 | RNAseq | 0.3268 | 0.0689 | |
| SRP159526 | CCS | 9973 | RNAseq | 0.3887 | 0.2050 | |
| SRP193095 | CCS | 9973 | RNAseq | 0.1928 | 0.0499 | |
| SRP219564 | CCS | 9973 | RNAseq | 0.0894 | 0.8157 | |
| TCGA | CCS | 9973 | RNAseq | -0.0442 | 0.4798 |
Upregulated datasets: 0; Downregulated datasets: 0.
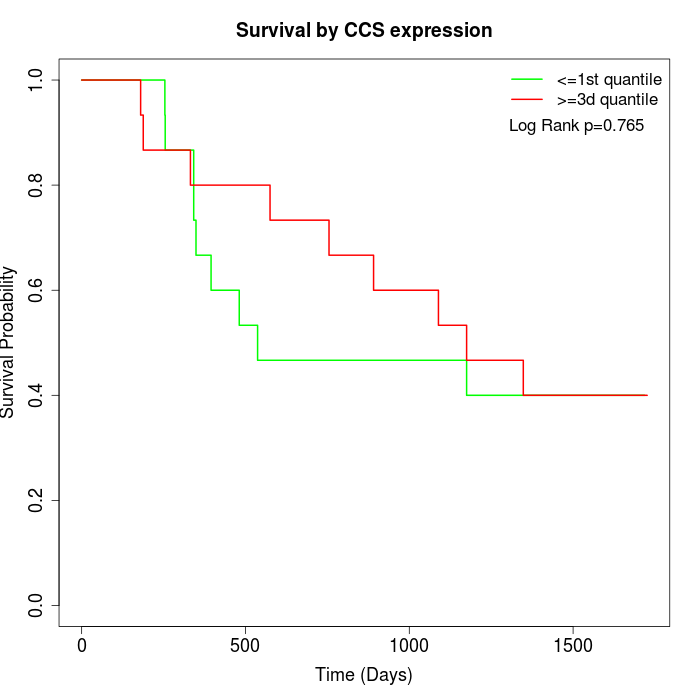
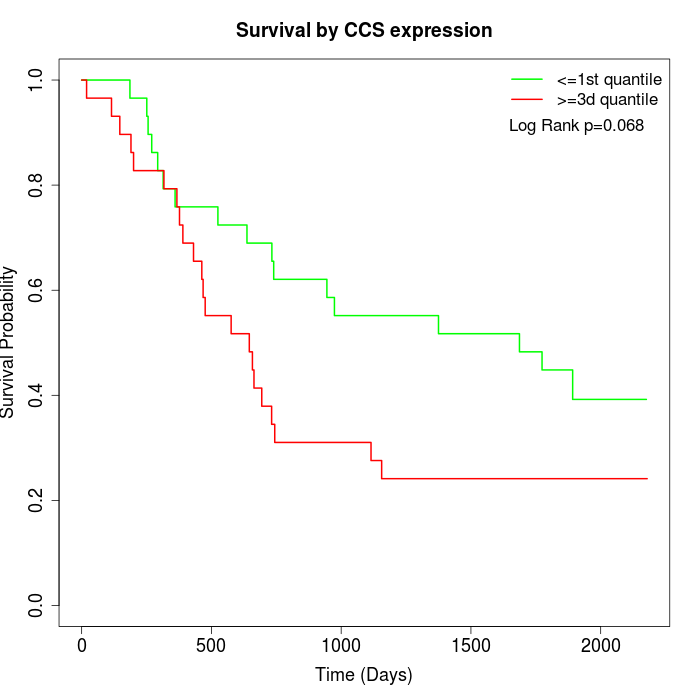
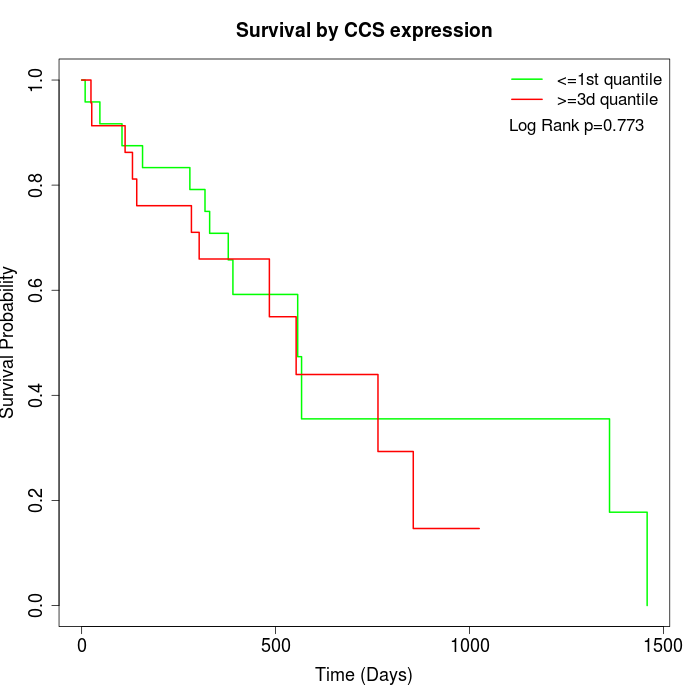
Survival by CCS expression:
Note: Click image to view full size file.
Copy number change of CCS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCS | 9973 | 9 | 4 | 17 | |
| GSE20123 | CCS | 9973 | 9 | 4 | 17 | |
| GSE43470 | CCS | 9973 | 6 | 1 | 36 | |
| GSE46452 | CCS | 9973 | 12 | 3 | 44 | |
| GSE47630 | CCS | 9973 | 9 | 4 | 27 | |
| GSE54993 | CCS | 9973 | 3 | 0 | 67 | |
| GSE54994 | CCS | 9973 | 9 | 5 | 39 | |
| GSE60625 | CCS | 9973 | 0 | 3 | 8 | |
| GSE74703 | CCS | 9973 | 4 | 0 | 32 | |
| GSE74704 | CCS | 9973 | 7 | 2 | 11 | |
| TCGA | CCS | 9973 | 27 | 6 | 63 |
Total number of gains: 95; Total number of losses: 32; Total Number of normals: 361.
Somatic mutations of CCS:
Generating mutation plots.
Highly correlated genes for CCS:
Showing top 20/583 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCS | CCDC127 | 0.84832 | 3 | 0 | 3 |
| CCS | ARFRP1 | 0.826278 | 3 | 0 | 3 |
| CCS | PPP1R16A | 0.823566 | 3 | 0 | 3 |
| CCS | WDR5 | 0.785857 | 3 | 0 | 3 |
| CCS | TP53I13 | 0.777651 | 3 | 0 | 3 |
| CCS | RP9 | 0.774096 | 3 | 0 | 3 |
| CCS | NAP1L4 | 0.755408 | 3 | 0 | 3 |
| CCS | UTP23 | 0.750633 | 4 | 0 | 4 |
| CCS | MED29 | 0.748189 | 3 | 0 | 3 |
| CCS | NGF | 0.745632 | 3 | 0 | 3 |
| CCS | MGAT5 | 0.739962 | 3 | 0 | 3 |
| CCS | HDAC11 | 0.739093 | 3 | 0 | 3 |
| CCS | BLCAP | 0.734222 | 4 | 0 | 4 |
| CCS | IL17RD | 0.728137 | 4 | 0 | 3 |
| CCS | SLC27A3 | 0.724169 | 3 | 0 | 3 |
| CCS | PKNOX1 | 0.719453 | 5 | 0 | 4 |
| CCS | SMCHD1 | 0.716529 | 3 | 0 | 3 |
| CCS | OLA1 | 0.713768 | 3 | 0 | 3 |
| CCS | CRTC2 | 0.712973 | 3 | 0 | 3 |
| CCS | KHNYN | 0.712838 | 3 | 0 | 3 |
For details and further investigation, click here