| Full name: cyclin dependent kinase 2 interacting protein | Alias Symbol: MGC849 | ||
| Type: protein-coding gene | Cytoband: 14q32.31 | ||
| Entrez ID: 51550 | HGNC ID: HGNC:23789 | Ensembl Gene: ENSG00000100865 | OMIM ID: 613362 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CINP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CINP | 51550 | 218267_at | 0.1764 | 0.7215 | |
| GSE20347 | CINP | 51550 | 218267_at | -0.0287 | 0.8487 | |
| GSE23400 | CINP | 51550 | 218267_at | 0.0456 | 0.4608 | |
| GSE26886 | CINP | 51550 | 218267_at | -0.1176 | 0.5062 | |
| GSE29001 | CINP | 51550 | 217598_at | -0.0660 | 0.8052 | |
| GSE38129 | CINP | 51550 | 218267_at | 0.0838 | 0.5038 | |
| GSE45670 | CINP | 51550 | 218267_at | 0.1383 | 0.3648 | |
| GSE53622 | CINP | 51550 | 67840 | 0.1618 | 0.0440 | |
| GSE53624 | CINP | 51550 | 67840 | 0.0672 | 0.4176 | |
| GSE63941 | CINP | 51550 | 218267_at | 0.0793 | 0.8860 | |
| GSE77861 | CINP | 51550 | 218267_at | -0.1048 | 0.6893 | |
| GSE97050 | CINP | 51550 | A_23_P88134 | 0.1155 | 0.6509 | |
| SRP007169 | CINP | 51550 | RNAseq | 0.2133 | 0.6509 | |
| SRP008496 | CINP | 51550 | RNAseq | 0.2659 | 0.4976 | |
| SRP064894 | CINP | 51550 | RNAseq | 0.3405 | 0.1693 | |
| SRP133303 | CINP | 51550 | RNAseq | 0.2635 | 0.0680 | |
| SRP159526 | CINP | 51550 | RNAseq | 0.2385 | 0.5855 | |
| SRP193095 | CINP | 51550 | RNAseq | -0.1988 | 0.2220 | |
| SRP219564 | CINP | 51550 | RNAseq | 0.0890 | 0.7987 | |
| TCGA | CINP | 51550 | RNAseq | 0.0390 | 0.5393 |
Upregulated datasets: 0; Downregulated datasets: 0.
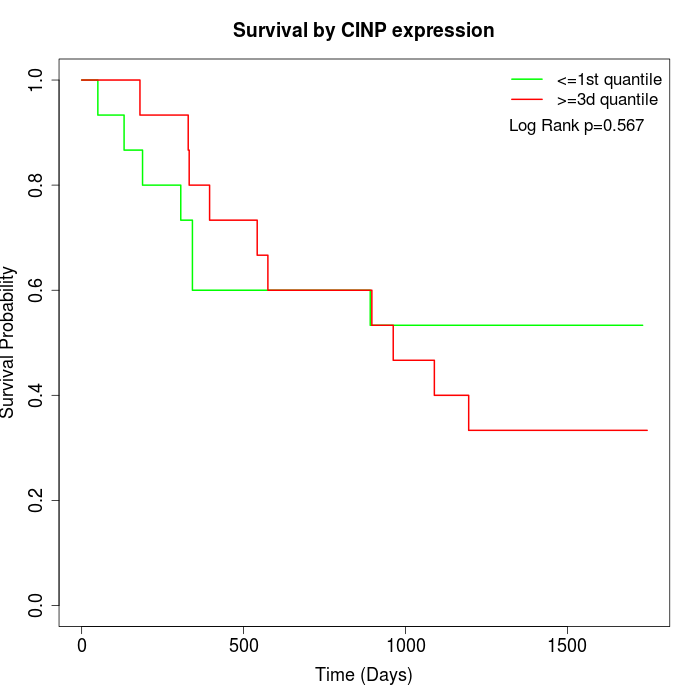
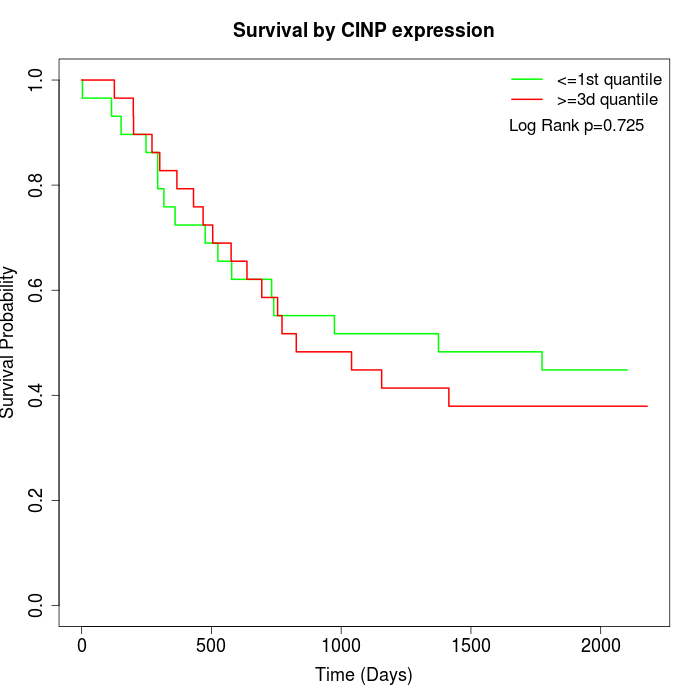
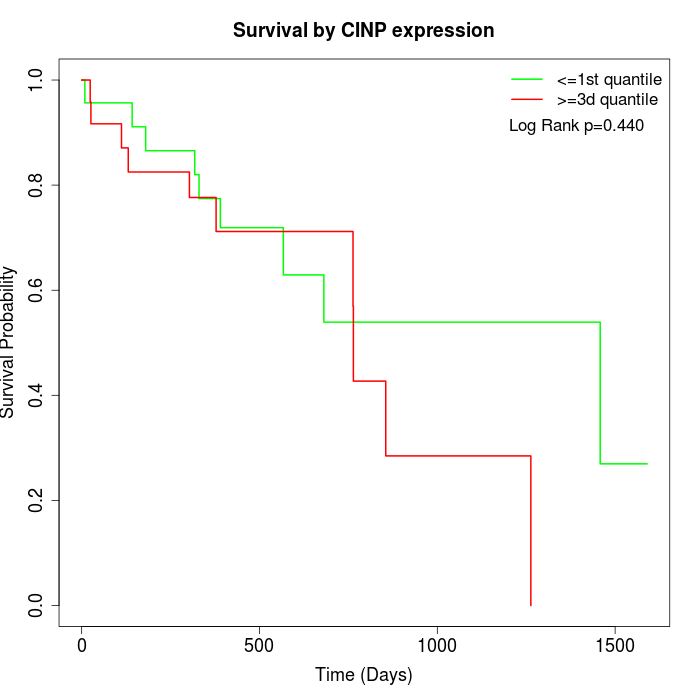
Survival by CINP expression:
Note: Click image to view full size file.
Copy number change of CINP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CINP | 51550 | 11 | 5 | 14 | |
| GSE20123 | CINP | 51550 | 10 | 4 | 16 | |
| GSE43470 | CINP | 51550 | 7 | 4 | 32 | |
| GSE46452 | CINP | 51550 | 18 | 4 | 37 | |
| GSE47630 | CINP | 51550 | 10 | 9 | 21 | |
| GSE54993 | CINP | 51550 | 3 | 8 | 59 | |
| GSE54994 | CINP | 51550 | 21 | 4 | 28 | |
| GSE60625 | CINP | 51550 | 0 | 2 | 9 | |
| GSE74703 | CINP | 51550 | 6 | 3 | 27 | |
| GSE74704 | CINP | 51550 | 5 | 3 | 12 | |
| TCGA | CINP | 51550 | 32 | 19 | 45 |
Total number of gains: 123; Total number of losses: 65; Total Number of normals: 300.
Somatic mutations of CINP:
Generating mutation plots.
Highly correlated genes for CINP:
Showing top 20/194 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CINP | RFC1 | 0.723936 | 3 | 0 | 3 |
| CINP | PODNL1 | 0.700117 | 3 | 0 | 3 |
| CINP | PIGF | 0.692726 | 4 | 0 | 4 |
| CINP | NOXO1 | 0.692457 | 3 | 0 | 3 |
| CINP | CFDP1 | 0.679775 | 3 | 0 | 3 |
| CINP | KLHDC2 | 0.678759 | 3 | 0 | 3 |
| CINP | AP4E1 | 0.67759 | 3 | 0 | 3 |
| CINP | TMTC3 | 0.677126 | 3 | 0 | 3 |
| CINP | DCLRE1B | 0.670166 | 3 | 0 | 3 |
| CINP | ZNF70 | 0.667464 | 3 | 0 | 3 |
| CINP | PTGR1 | 0.665321 | 3 | 0 | 3 |
| CINP | TK2 | 0.654216 | 3 | 0 | 3 |
| CINP | PRELID1 | 0.653341 | 4 | 0 | 4 |
| CINP | WDR20 | 0.650399 | 6 | 0 | 5 |
| CINP | HKDC1 | 0.650383 | 4 | 0 | 3 |
| CINP | COPS8 | 0.639058 | 3 | 0 | 3 |
| CINP | PGLYRP4 | 0.636658 | 3 | 0 | 3 |
| CINP | CEP76 | 0.634504 | 4 | 0 | 3 |
| CINP | FDPS | 0.63314 | 5 | 0 | 3 |
| CINP | TTC23 | 0.630722 | 3 | 0 | 3 |
For details and further investigation, click here