| Full name: chloride channel accessory 2 | Alias Symbol: CLCRG2 | ||
| Type: protein-coding gene | Cytoband: 1p22.3 | ||
| Entrez ID: 9635 | HGNC ID: HGNC:2016 | Ensembl Gene: ENSG00000137975 | OMIM ID: 604003 |
| Drug and gene relationship at DGIdb | |||
CLCA2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04972 | Pancreatic secretion |
Expression of CLCA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLCA2 | 9635 | 206165_s_at | -0.0422 | 0.9828 | |
| GSE20347 | CLCA2 | 9635 | 206165_s_at | -1.4611 | 0.0004 | |
| GSE23400 | CLCA2 | 9635 | 206165_s_at | -0.2433 | 0.2734 | |
| GSE26886 | CLCA2 | 9635 | 206165_s_at | -2.1449 | 0.0010 | |
| GSE29001 | CLCA2 | 9635 | 206165_s_at | -0.7342 | 0.3303 | |
| GSE38129 | CLCA2 | 9635 | 206165_s_at | 0.1869 | 0.8269 | |
| GSE45670 | CLCA2 | 9635 | 206165_s_at | 0.3805 | 0.0830 | |
| GSE53622 | CLCA2 | 9635 | 122789 | -0.1692 | 0.6233 | |
| GSE53624 | CLCA2 | 9635 | 122789 | -0.6711 | 0.0001 | |
| GSE63941 | CLCA2 | 9635 | 206165_s_at | 1.1245 | 0.5917 | |
| GSE77861 | CLCA2 | 9635 | 206165_s_at | -0.3277 | 0.1267 | |
| GSE97050 | CLCA2 | 9635 | A_23_P397248 | 0.2983 | 0.7972 | |
| SRP007169 | CLCA2 | 9635 | RNAseq | -1.9630 | 0.0004 | |
| SRP008496 | CLCA2 | 9635 | RNAseq | -1.4340 | 0.0001 | |
| SRP064894 | CLCA2 | 9635 | RNAseq | -0.0918 | 0.8440 | |
| SRP133303 | CLCA2 | 9635 | RNAseq | -0.2364 | 0.4968 | |
| SRP159526 | CLCA2 | 9635 | RNAseq | -0.8166 | 0.2023 | |
| SRP219564 | CLCA2 | 9635 | RNAseq | -0.2257 | 0.8700 | |
| TCGA | CLCA2 | 9635 | RNAseq | 1.4867 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 4.
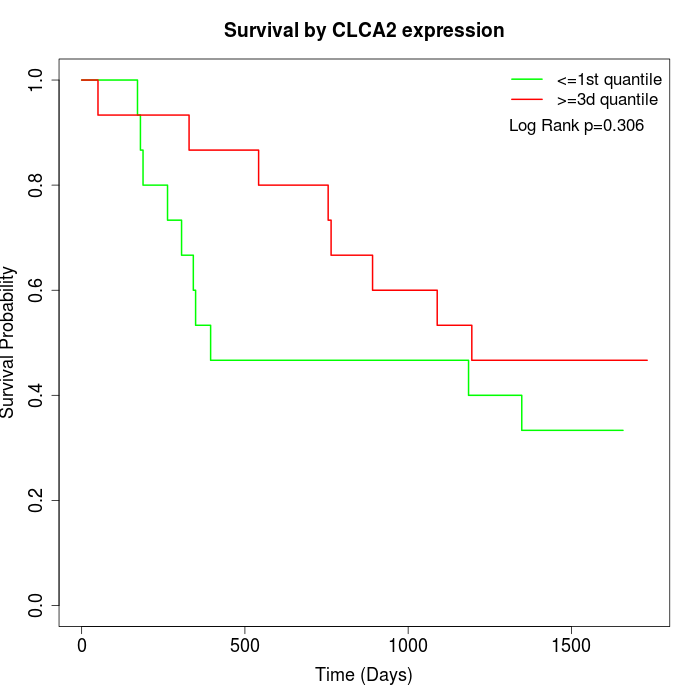
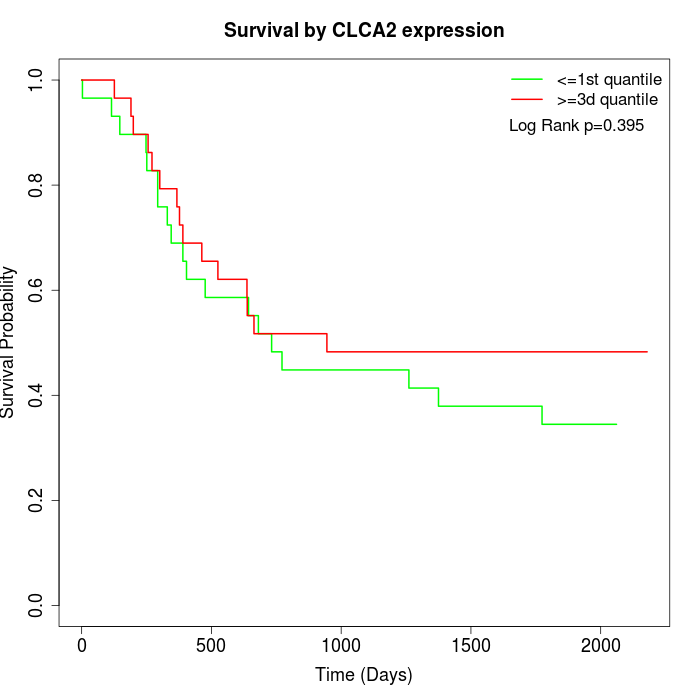
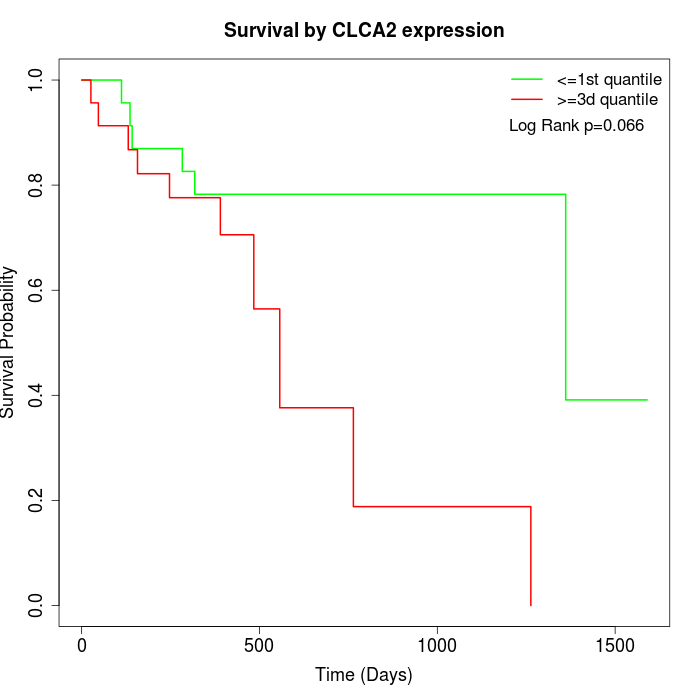
Survival by CLCA2 expression:
Note: Click image to view full size file.
Copy number change of CLCA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLCA2 | 9635 | 0 | 8 | 22 | |
| GSE20123 | CLCA2 | 9635 | 0 | 8 | 22 | |
| GSE43470 | CLCA2 | 9635 | 2 | 2 | 39 | |
| GSE46452 | CLCA2 | 9635 | 1 | 1 | 57 | |
| GSE47630 | CLCA2 | 9635 | 8 | 5 | 27 | |
| GSE54993 | CLCA2 | 9635 | 0 | 1 | 69 | |
| GSE54994 | CLCA2 | 9635 | 6 | 3 | 44 | |
| GSE60625 | CLCA2 | 9635 | 0 | 0 | 11 | |
| GSE74703 | CLCA2 | 9635 | 1 | 2 | 33 | |
| GSE74704 | CLCA2 | 9635 | 0 | 5 | 15 | |
| TCGA | CLCA2 | 9635 | 6 | 23 | 67 |
Total number of gains: 24; Total number of losses: 58; Total Number of normals: 406.
Somatic mutations of CLCA2:
Generating mutation plots.
Highly correlated genes for CLCA2:
Showing top 20/570 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLCA2 | ARAP2 | 0.742046 | 3 | 0 | 3 |
| CLCA2 | SERPINB5 | 0.716486 | 11 | 0 | 10 |
| CLCA2 | KRT6A | 0.711357 | 12 | 0 | 10 |
| CLCA2 | KRT6C | 0.711007 | 4 | 0 | 3 |
| CLCA2 | KRT6B | 0.69838 | 12 | 0 | 10 |
| CLCA2 | DSG3 | 0.695185 | 12 | 0 | 10 |
| CLCA2 | DMKN | 0.69364 | 6 | 0 | 5 |
| CLCA2 | TMEM79 | 0.690889 | 7 | 0 | 7 |
| CLCA2 | PKP1 | 0.690396 | 11 | 0 | 9 |
| CLCA2 | S100A11 | 0.681096 | 9 | 0 | 7 |
| CLCA2 | ZCCHC9 | 0.679406 | 3 | 0 | 3 |
| CLCA2 | KRT80 | 0.675266 | 5 | 0 | 5 |
| CLCA2 | TRIM29 | 0.672267 | 10 | 0 | 8 |
| CLCA2 | FABP5 | 0.670063 | 11 | 0 | 9 |
| CLCA2 | GSDMC | 0.66887 | 8 | 0 | 7 |
| CLCA2 | SLC25A43 | 0.66662 | 4 | 0 | 4 |
| CLCA2 | IFI27L1 | 0.665172 | 3 | 0 | 3 |
| CLCA2 | GJB6 | 0.665088 | 9 | 0 | 7 |
| CLCA2 | LAD1 | 0.66142 | 10 | 0 | 8 |
| CLCA2 | GNA15 | 0.66013 | 13 | 0 | 12 |
For details and further investigation, click here