| Full name: G protein subunit alpha 15 | Alias Symbol: GNA16 | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 2769 | HGNC ID: HGNC:4383 | Ensembl Gene: ENSG00000060558 | OMIM ID: 139314 |
| Drug and gene relationship at DGIdb | |||
GNA15 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa05142 | Chagas disease (American trypanosomiasis) |
Expression of GNA15:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GNA15 | 2769 | 205349_at | -0.1892 | 0.8783 | |
| GSE20347 | GNA15 | 2769 | 205349_at | -1.2769 | 0.0000 | |
| GSE23400 | GNA15 | 2769 | 205349_at | -0.7709 | 0.0000 | |
| GSE26886 | GNA15 | 2769 | 205349_at | -1.2608 | 0.0002 | |
| GSE29001 | GNA15 | 2769 | 205349_at | -1.2361 | 0.0016 | |
| GSE38129 | GNA15 | 2769 | 205349_at | -0.4505 | 0.3580 | |
| GSE45670 | GNA15 | 2769 | 205349_at | 0.0177 | 0.9395 | |
| GSE53622 | GNA15 | 2769 | 27742 | -0.6775 | 0.0048 | |
| GSE53624 | GNA15 | 2769 | 27742 | -1.0003 | 0.0000 | |
| GSE63941 | GNA15 | 2769 | 205349_at | 2.7545 | 0.0076 | |
| GSE77861 | GNA15 | 2769 | 205349_at | -0.5581 | 0.0079 | |
| GSE97050 | GNA15 | 2769 | A_24_P331128 | 0.5459 | 0.7069 | |
| SRP007169 | GNA15 | 2769 | RNAseq | -2.7973 | 0.0000 | |
| SRP008496 | GNA15 | 2769 | RNAseq | -2.5290 | 0.0000 | |
| SRP064894 | GNA15 | 2769 | RNAseq | -1.4097 | 0.0000 | |
| SRP133303 | GNA15 | 2769 | RNAseq | -0.9026 | 0.0003 | |
| SRP159526 | GNA15 | 2769 | RNAseq | -1.8608 | 0.0169 | |
| SRP193095 | GNA15 | 2769 | RNAseq | -1.2399 | 0.0008 | |
| SRP219564 | GNA15 | 2769 | RNAseq | -0.8863 | 0.3088 | |
| TCGA | GNA15 | 2769 | RNAseq | 0.8436 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 9.
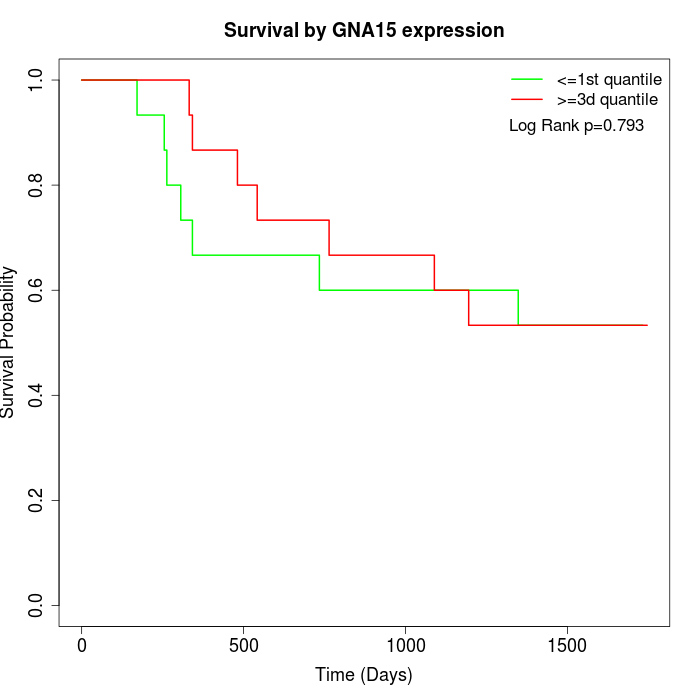
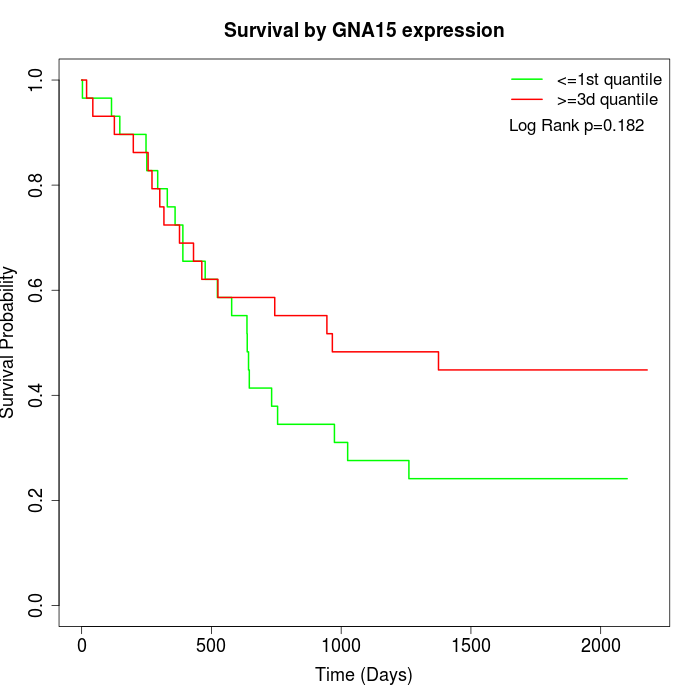
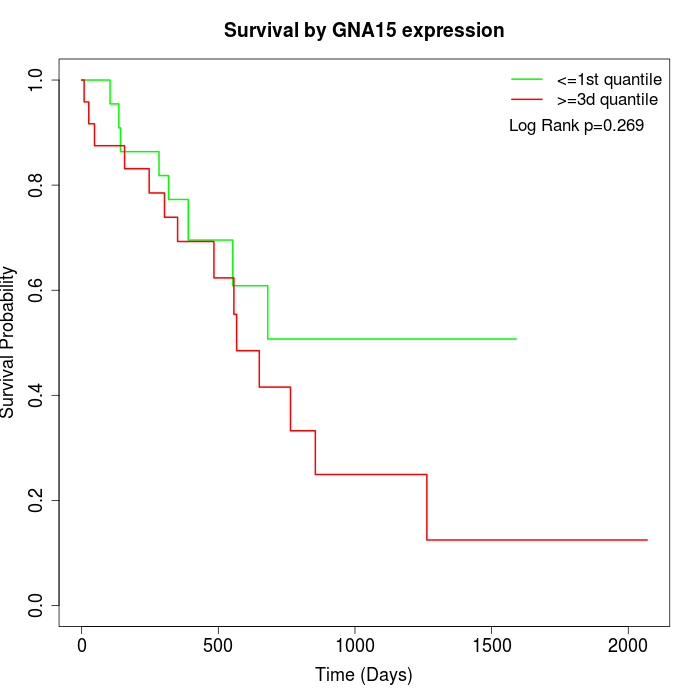
Survival by GNA15 expression:
Note: Click image to view full size file.
Copy number change of GNA15:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GNA15 | 2769 | 4 | 4 | 22 | |
| GSE20123 | GNA15 | 2769 | 3 | 4 | 23 | |
| GSE43470 | GNA15 | 2769 | 1 | 9 | 33 | |
| GSE46452 | GNA15 | 2769 | 47 | 1 | 11 | |
| GSE47630 | GNA15 | 2769 | 5 | 7 | 28 | |
| GSE54993 | GNA15 | 2769 | 16 | 3 | 51 | |
| GSE54994 | GNA15 | 2769 | 8 | 16 | 29 | |
| GSE60625 | GNA15 | 2769 | 9 | 0 | 2 | |
| GSE74703 | GNA15 | 2769 | 1 | 6 | 29 | |
| GSE74704 | GNA15 | 2769 | 1 | 3 | 16 | |
| TCGA | GNA15 | 2769 | 8 | 22 | 66 |
Total number of gains: 103; Total number of losses: 75; Total Number of normals: 310.
Somatic mutations of GNA15:
Generating mutation plots.
Highly correlated genes for GNA15:
Showing top 20/1136 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GNA15 | KRT6C | 0.796602 | 4 | 0 | 4 |
| GNA15 | LYPD3 | 0.776244 | 13 | 0 | 13 |
| GNA15 | PKP3 | 0.768815 | 13 | 0 | 12 |
| GNA15 | S100A11 | 0.760786 | 12 | 0 | 12 |
| GNA15 | SH3GL1 | 0.754996 | 11 | 0 | 11 |
| GNA15 | LAD1 | 0.744439 | 13 | 0 | 12 |
| GNA15 | GJB2 | 0.743698 | 7 | 0 | 7 |
| GNA15 | DSG3 | 0.73929 | 13 | 0 | 12 |
| GNA15 | TMEM40 | 0.736431 | 13 | 0 | 13 |
| GNA15 | PPP1R13L | 0.735592 | 13 | 0 | 13 |
| GNA15 | PGM2 | 0.735477 | 7 | 0 | 7 |
| GNA15 | MALL | 0.734896 | 13 | 0 | 13 |
| GNA15 | S100A14 | 0.734758 | 13 | 0 | 13 |
| GNA15 | TMEM154 | 0.730984 | 8 | 0 | 8 |
| GNA15 | JUP | 0.730435 | 12 | 0 | 11 |
| GNA15 | S100A16 | 0.727171 | 9 | 0 | 8 |
| GNA15 | RHOD | 0.726487 | 13 | 0 | 12 |
| GNA15 | C1orf116 | 0.726399 | 13 | 0 | 13 |
| GNA15 | ANXA2 | 0.72636 | 12 | 0 | 11 |
| GNA15 | UPP1 | 0.721206 | 12 | 0 | 11 |
For details and further investigation, click here