| Full name: chloride voltage-gated channel 1 | Alias Symbol: CLC1|ClC-1 | ||
| Type: protein-coding gene | Cytoband: 7q34 | ||
| Entrez ID: 1180 | HGNC ID: HGNC:2019 | Ensembl Gene: ENSG00000188037 | OMIM ID: 118425 |
| Drug and gene relationship at DGIdb | |||
Expression of CLCN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLCN1 | 1180 | 208437_at | -0.1565 | 0.5827 | |
| GSE20347 | CLCN1 | 1180 | 208437_at | 0.0437 | 0.6094 | |
| GSE23400 | CLCN1 | 1180 | 208437_at | -0.0911 | 0.0069 | |
| GSE26886 | CLCN1 | 1180 | 208437_at | -0.0337 | 0.8292 | |
| GSE29001 | CLCN1 | 1180 | 208437_at | -0.2793 | 0.0333 | |
| GSE38129 | CLCN1 | 1180 | 208437_at | -0.0604 | 0.5000 | |
| GSE45670 | CLCN1 | 1180 | 208437_at | 0.0105 | 0.9327 | |
| GSE53622 | CLCN1 | 1180 | 37656 | -0.2009 | 0.0684 | |
| GSE53624 | CLCN1 | 1180 | 37656 | -0.4288 | 0.0000 | |
| GSE63941 | CLCN1 | 1180 | 208437_at | 0.0445 | 0.8700 | |
| GSE77861 | CLCN1 | 1180 | 208437_at | -0.1284 | 0.2783 | |
| GSE97050 | CLCN1 | 1180 | A_23_P59772 | -0.0677 | 0.7829 | |
| SRP159526 | CLCN1 | 1180 | RNAseq | -0.6779 | 0.4193 | |
| TCGA | CLCN1 | 1180 | RNAseq | 0.5217 | 0.3851 |
Upregulated datasets: 0; Downregulated datasets: 0.
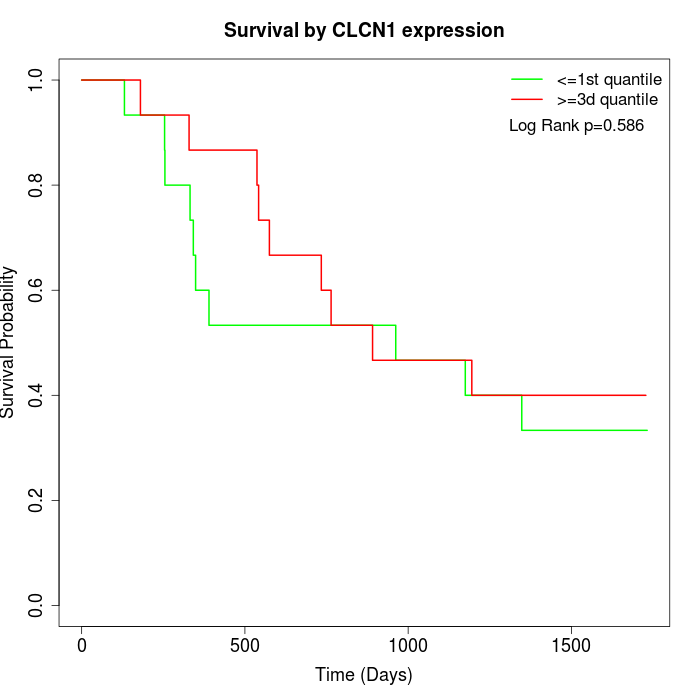
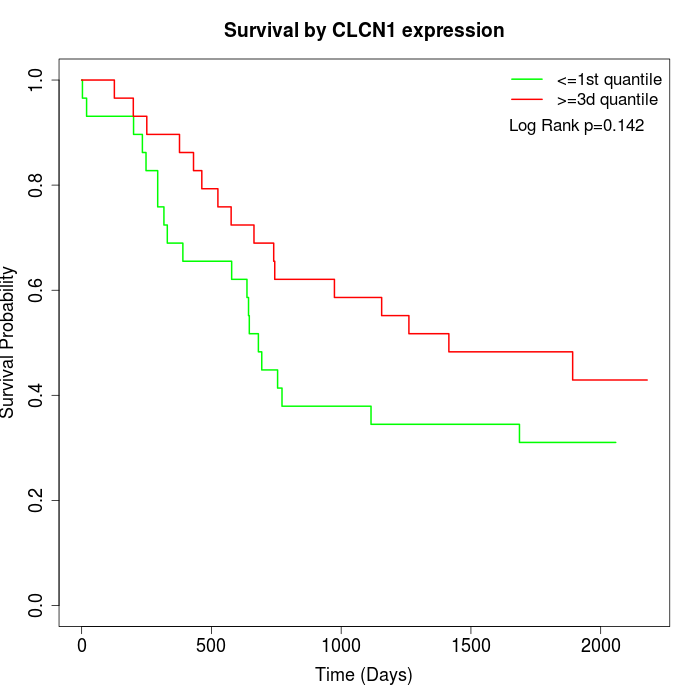
Survival by CLCN1 expression:
Note: Click image to view full size file.
Copy number change of CLCN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLCN1 | 1180 | 4 | 2 | 24 | |
| GSE20123 | CLCN1 | 1180 | 4 | 2 | 24 | |
| GSE43470 | CLCN1 | 1180 | 3 | 4 | 36 | |
| GSE46452 | CLCN1 | 1180 | 7 | 2 | 50 | |
| GSE47630 | CLCN1 | 1180 | 6 | 7 | 27 | |
| GSE54993 | CLCN1 | 1180 | 3 | 4 | 63 | |
| GSE54994 | CLCN1 | 1180 | 5 | 8 | 40 | |
| GSE60625 | CLCN1 | 1180 | 0 | 0 | 11 | |
| GSE74703 | CLCN1 | 1180 | 2 | 4 | 30 | |
| GSE74704 | CLCN1 | 1180 | 2 | 2 | 16 | |
| TCGA | CLCN1 | 1180 | 28 | 25 | 43 |
Total number of gains: 64; Total number of losses: 60; Total Number of normals: 364.
Somatic mutations of CLCN1:
Generating mutation plots.
Highly correlated genes for CLCN1:
Showing top 20/524 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLCN1 | PTGDR2 | 0.793136 | 3 | 0 | 3 |
| CLCN1 | ARHGAP30 | 0.77479 | 3 | 0 | 3 |
| CLCN1 | CDX2 | 0.730112 | 5 | 0 | 5 |
| CLCN1 | ENHO | 0.728497 | 3 | 0 | 3 |
| CLCN1 | PRG2 | 0.727825 | 3 | 0 | 3 |
| CLCN1 | RASL10B | 0.723866 | 3 | 0 | 3 |
| CLCN1 | TMEM37 | 0.721746 | 3 | 0 | 3 |
| CLCN1 | KRT74 | 0.721166 | 3 | 0 | 3 |
| CLCN1 | NKX6-3 | 0.719396 | 3 | 0 | 3 |
| CLCN1 | CPNE9 | 0.718208 | 3 | 0 | 3 |
| CLCN1 | LRFN3 | 0.704944 | 3 | 0 | 3 |
| CLCN1 | ST8SIA3 | 0.701295 | 3 | 0 | 3 |
| CLCN1 | ATP8B3 | 0.700743 | 3 | 0 | 3 |
| CLCN1 | NRG2 | 0.695154 | 3 | 0 | 3 |
| CLCN1 | CCL25 | 0.686207 | 4 | 0 | 3 |
| CLCN1 | EXOSC1 | 0.680552 | 3 | 0 | 3 |
| CLCN1 | PGM5 | 0.678969 | 3 | 0 | 3 |
| CLCN1 | NKAIN4 | 0.678715 | 3 | 0 | 3 |
| CLCN1 | CNGB1 | 0.677947 | 6 | 0 | 5 |
| CLCN1 | OR2C3 | 0.676706 | 3 | 0 | 3 |
For details and further investigation, click here