| Full name: chloride voltage-gated channel 5 | Alias Symbol: DENTS|XLRH|hClC-K2|hCIC-K2|CLC5|XRN|ClC-5 | ||
| Type: protein-coding gene | Cytoband: Xp11.23 | ||
| Entrez ID: 1184 | HGNC ID: HGNC:2023 | Ensembl Gene: ENSG00000171365 | OMIM ID: 300008 |
| Drug and gene relationship at DGIdb | |||
Expression of CLCN5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLCN5 | 1184 | 226274_at | 0.4648 | 0.4124 | |
| GSE20347 | CLCN5 | 1184 | 206704_at | 0.0892 | 0.3577 | |
| GSE23400 | CLCN5 | 1184 | 206704_at | -0.0290 | 0.3377 | |
| GSE26886 | CLCN5 | 1184 | 226274_at | 0.2027 | 0.4483 | |
| GSE29001 | CLCN5 | 1184 | 206704_at | -0.1250 | 0.4178 | |
| GSE38129 | CLCN5 | 1184 | 206704_at | 0.2747 | 0.1130 | |
| GSE45670 | CLCN5 | 1184 | 226274_at | 0.2822 | 0.1022 | |
| GSE53622 | CLCN5 | 1184 | 27904 | -0.2556 | 0.0216 | |
| GSE53624 | CLCN5 | 1184 | 27904 | 0.0933 | 0.3184 | |
| GSE63941 | CLCN5 | 1184 | 226274_at | -0.6510 | 0.2062 | |
| GSE77861 | CLCN5 | 1184 | 226274_at | 0.1838 | 0.3681 | |
| GSE97050 | CLCN5 | 1184 | A_33_P3415087 | 0.3853 | 0.1470 | |
| SRP007169 | CLCN5 | 1184 | RNAseq | 0.7982 | 0.0520 | |
| SRP008496 | CLCN5 | 1184 | RNAseq | 0.7495 | 0.0397 | |
| SRP064894 | CLCN5 | 1184 | RNAseq | 0.0921 | 0.6992 | |
| SRP133303 | CLCN5 | 1184 | RNAseq | -0.1442 | 0.5098 | |
| SRP159526 | CLCN5 | 1184 | RNAseq | 0.2171 | 0.3912 | |
| SRP193095 | CLCN5 | 1184 | RNAseq | 0.0265 | 0.8641 | |
| SRP219564 | CLCN5 | 1184 | RNAseq | 0.2770 | 0.5858 | |
| TCGA | CLCN5 | 1184 | RNAseq | -0.2237 | 0.0064 |
Upregulated datasets: 0; Downregulated datasets: 0.
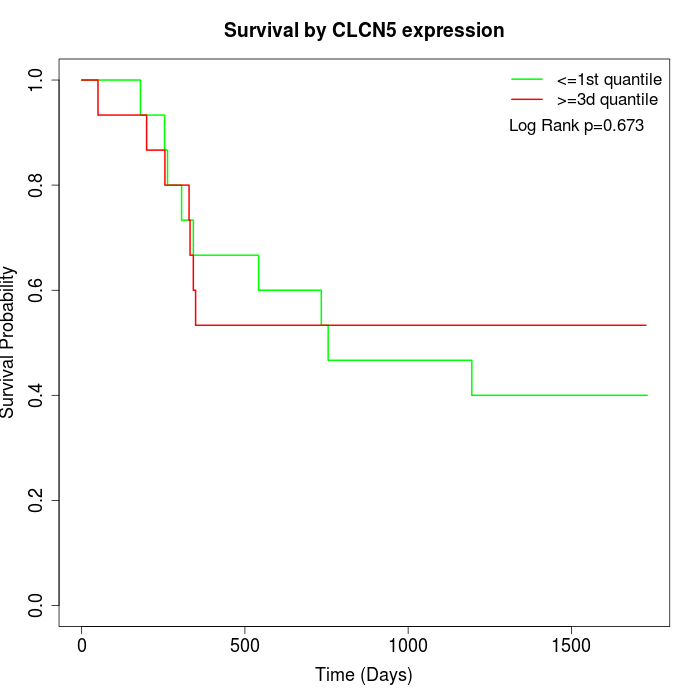
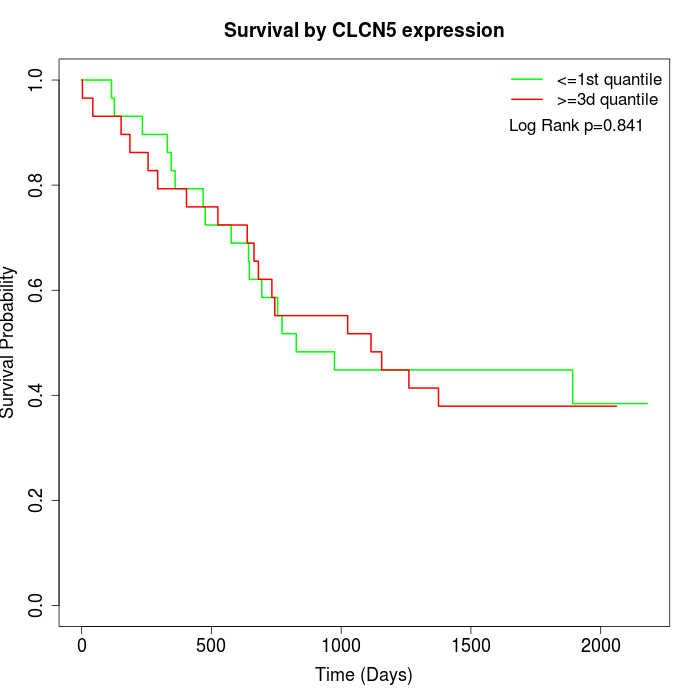
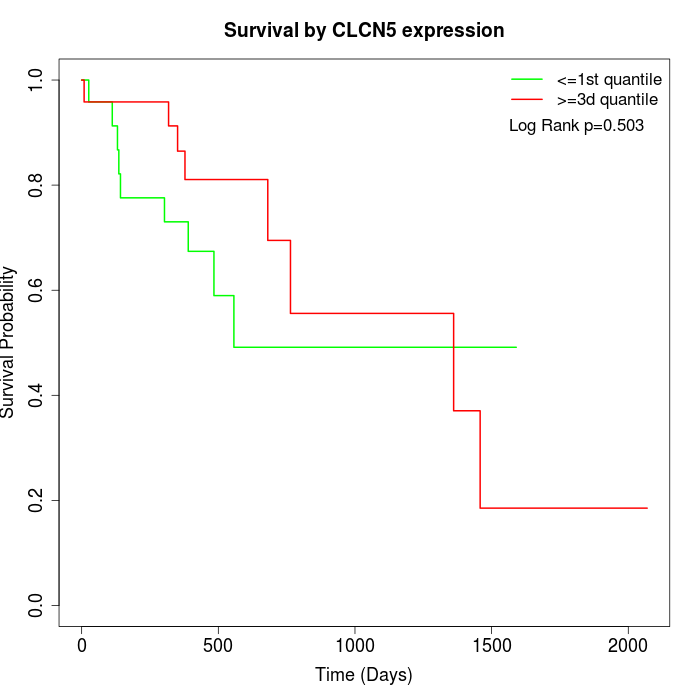
Survival by CLCN5 expression:
Note: Click image to view full size file.
Copy number change of CLCN5:
No record found for this gene.
Somatic mutations of CLCN5:
Generating mutation plots.
Highly correlated genes for CLCN5:
Showing top 20/151 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLCN5 | RPP38 | 0.771058 | 3 | 0 | 3 |
| CLCN5 | BUD31 | 0.756698 | 3 | 0 | 3 |
| CLCN5 | ANKIB1 | 0.739093 | 3 | 0 | 3 |
| CLCN5 | SIX4 | 0.736067 | 3 | 0 | 3 |
| CLCN5 | WDR3 | 0.712686 | 3 | 0 | 3 |
| CLCN5 | NIPA2 | 0.709682 | 3 | 0 | 3 |
| CLCN5 | SLC30A6 | 0.705596 | 3 | 0 | 3 |
| CLCN5 | PPP1R35 | 0.695839 | 3 | 0 | 3 |
| CLCN5 | ALDH5A1 | 0.689477 | 4 | 0 | 3 |
| CLCN5 | SHMT2 | 0.686217 | 4 | 0 | 3 |
| CLCN5 | PRDM15 | 0.681606 | 3 | 0 | 3 |
| CLCN5 | STEAP1 | 0.670847 | 4 | 0 | 3 |
| CLCN5 | STK11IP | 0.668159 | 3 | 0 | 3 |
| CLCN5 | TXNDC12 | 0.667605 | 3 | 0 | 3 |
| CLCN5 | PEX10 | 0.665546 | 3 | 0 | 3 |
| CLCN5 | PUSL1 | 0.664651 | 3 | 0 | 3 |
| CLCN5 | TNFSF15 | 0.664555 | 3 | 0 | 3 |
| CLCN5 | PSTPIP2 | 0.663905 | 3 | 0 | 3 |
| CLCN5 | TOP3B | 0.656828 | 3 | 0 | 3 |
| CLCN5 | ANKRD54 | 0.654145 | 3 | 0 | 3 |
For details and further investigation, click here