| Full name: casein kinase 2 alpha 1 | Alias Symbol: Cka1|Cka2 | ||
| Type: protein-coding gene | Cytoband: 20p13 | ||
| Entrez ID: 1457 | HGNC ID: HGNC:2457 | Ensembl Gene: ENSG00000101266 | OMIM ID: 115440 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CSNK2A1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04064 | NF-kappa B signaling pathway | |
| hsa04310 | Wnt signaling pathway | |
| hsa04520 | Adherens junction | |
| hsa04530 | Tight junction | |
| hsa05168 | Herpes simplex infection |
Expression of CSNK2A1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CSNK2A1 | 1457 | 212072_s_at | 0.3121 | 0.5696 | |
| GSE20347 | CSNK2A1 | 1457 | 212072_s_at | 0.0463 | 0.7670 | |
| GSE23400 | CSNK2A1 | 1457 | 212072_s_at | 0.1420 | 0.0540 | |
| GSE26886 | CSNK2A1 | 1457 | 212072_s_at | -0.0452 | 0.8397 | |
| GSE29001 | CSNK2A1 | 1457 | 212072_s_at | 0.0997 | 0.5504 | |
| GSE38129 | CSNK2A1 | 1457 | 212072_s_at | 0.3063 | 0.0430 | |
| GSE45670 | CSNK2A1 | 1457 | 212072_s_at | 0.3591 | 0.0203 | |
| GSE53622 | CSNK2A1 | 1457 | 10034 | 0.2862 | 0.0000 | |
| GSE53624 | CSNK2A1 | 1457 | 10034 | 0.2678 | 0.0000 | |
| GSE63941 | CSNK2A1 | 1457 | 212072_s_at | 0.3204 | 0.5005 | |
| GSE77861 | CSNK2A1 | 1457 | 212072_s_at | 0.0457 | 0.8754 | |
| GSE97050 | CSNK2A1 | 1457 | A_23_P502575 | 0.2769 | 0.4981 | |
| SRP007169 | CSNK2A1 | 1457 | RNAseq | -0.8656 | 0.0190 | |
| SRP008496 | CSNK2A1 | 1457 | RNAseq | -0.5728 | 0.0143 | |
| SRP064894 | CSNK2A1 | 1457 | RNAseq | -0.0891 | 0.6884 | |
| SRP133303 | CSNK2A1 | 1457 | RNAseq | 0.2163 | 0.0430 | |
| SRP159526 | CSNK2A1 | 1457 | RNAseq | 0.2508 | 0.4636 | |
| SRP193095 | CSNK2A1 | 1457 | RNAseq | -0.0269 | 0.7430 | |
| SRP219564 | CSNK2A1 | 1457 | RNAseq | -0.2749 | 0.4147 | |
| TCGA | CSNK2A1 | 1457 | RNAseq | 0.0740 | 0.1113 |
Upregulated datasets: 0; Downregulated datasets: 0.
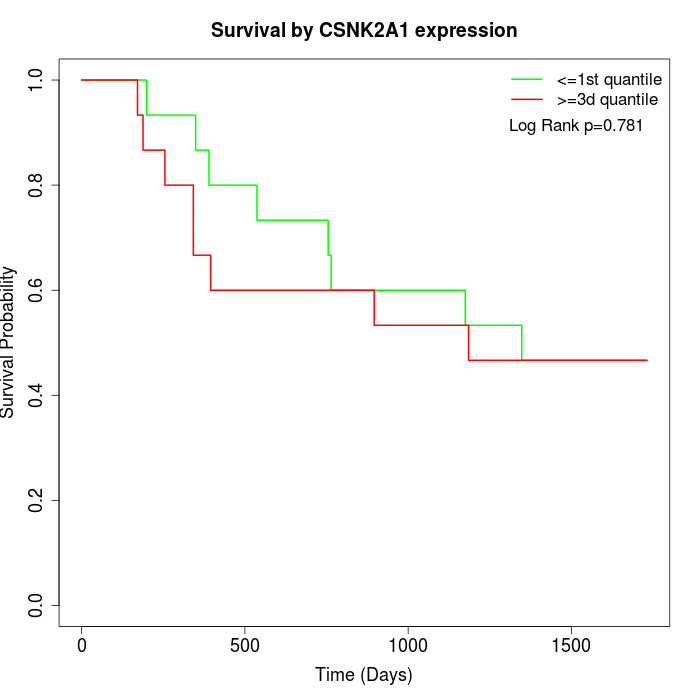
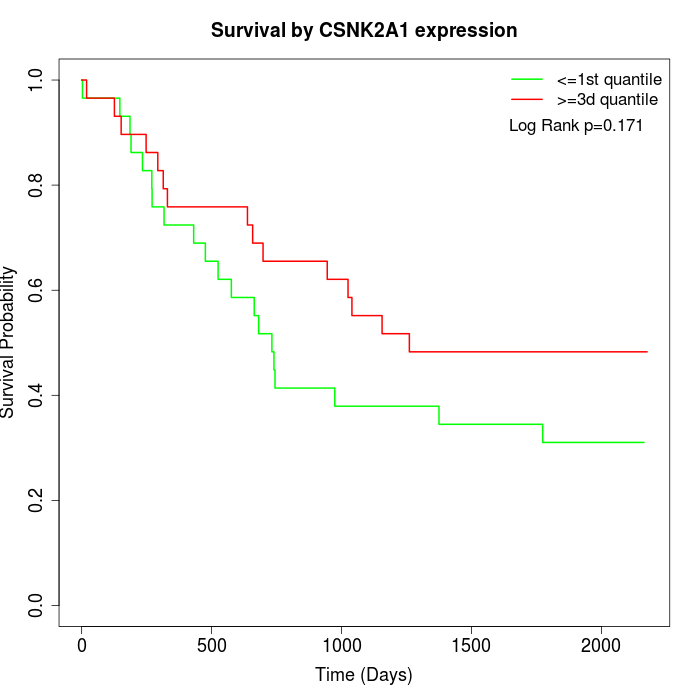
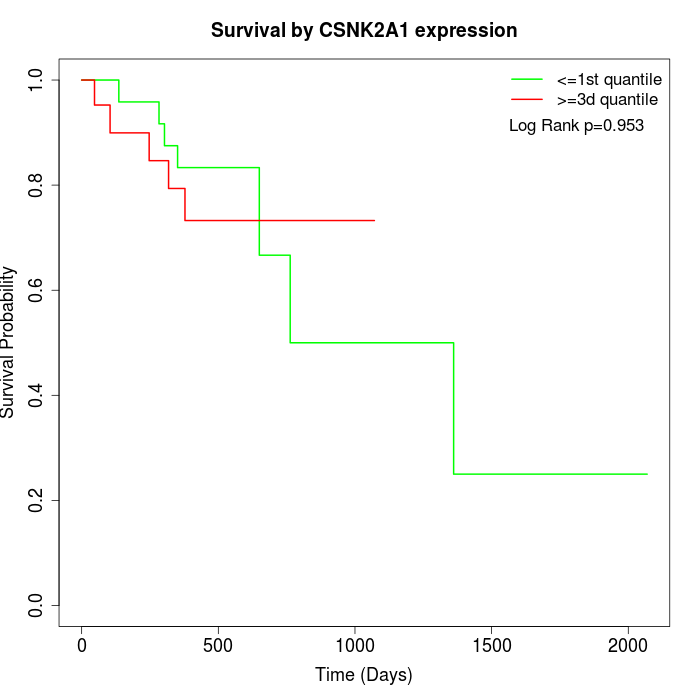
Survival by CSNK2A1 expression:
Note: Click image to view full size file.
Copy number change of CSNK2A1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CSNK2A1 | 1457 | 12 | 4 | 14 | |
| GSE20123 | CSNK2A1 | 1457 | 13 | 4 | 13 | |
| GSE43470 | CSNK2A1 | 1457 | 11 | 1 | 31 | |
| GSE46452 | CSNK2A1 | 1457 | 26 | 1 | 32 | |
| GSE47630 | CSNK2A1 | 1457 | 19 | 4 | 17 | |
| GSE54993 | CSNK2A1 | 1457 | 2 | 15 | 53 | |
| GSE54994 | CSNK2A1 | 1457 | 24 | 2 | 27 | |
| GSE60625 | CSNK2A1 | 1457 | 0 | 0 | 11 | |
| GSE74703 | CSNK2A1 | 1457 | 10 | 1 | 25 | |
| GSE74704 | CSNK2A1 | 1457 | 8 | 2 | 10 | |
| TCGA | CSNK2A1 | 1457 | 41 | 11 | 44 |
Total number of gains: 166; Total number of losses: 45; Total Number of normals: 277.
Somatic mutations of CSNK2A1:
Generating mutation plots.
Highly correlated genes for CSNK2A1:
Showing top 20/153 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CSNK2A1 | STK35 | 0.708722 | 7 | 0 | 6 |
| CSNK2A1 | LOR | 0.692707 | 4 | 0 | 3 |
| CSNK2A1 | TRMT6 | 0.678674 | 6 | 0 | 5 |
| CSNK2A1 | MBD1 | 0.673585 | 3 | 0 | 3 |
| CSNK2A1 | MTCH2 | 0.668875 | 3 | 0 | 3 |
| CSNK2A1 | FRMD8 | 0.667575 | 3 | 0 | 3 |
| CSNK2A1 | HDAC1 | 0.661807 | 4 | 0 | 3 |
| CSNK2A1 | ROBO1 | 0.65645 | 4 | 0 | 3 |
| CSNK2A1 | CBFB | 0.654852 | 3 | 0 | 3 |
| CSNK2A1 | NAA20 | 0.651019 | 5 | 0 | 3 |
| CSNK2A1 | UBA6 | 0.650898 | 4 | 0 | 3 |
| CSNK2A1 | FAM110A | 0.647123 | 6 | 0 | 5 |
| CSNK2A1 | GTF2IRD1 | 0.641226 | 4 | 0 | 3 |
| CSNK2A1 | RBCK1 | 0.636557 | 11 | 0 | 8 |
| CSNK2A1 | PET117 | 0.635886 | 5 | 0 | 4 |
| CSNK2A1 | GINS3 | 0.631767 | 4 | 0 | 3 |
| CSNK2A1 | TP53RK | 0.63131 | 3 | 0 | 3 |
| CSNK2A1 | NF2 | 0.626726 | 4 | 0 | 3 |
| CSNK2A1 | RALGAPA2 | 0.624567 | 5 | 0 | 5 |
| CSNK2A1 | NDUFV1 | 0.624402 | 3 | 0 | 3 |
For details and further investigation, click here