| Full name: loricrin | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1q21.3 | ||
| Entrez ID: 4014 | HGNC ID: HGNC:6663 | Ensembl Gene: ENSG00000203782 | OMIM ID: 152445 |
| Drug and gene relationship at DGIdb | |||
Expression of LOR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LOR | 4014 | 207720_at | 0.1818 | 0.8288 | |
| GSE20347 | LOR | 4014 | 207720_at | 0.5658 | 0.2697 | |
| GSE23400 | LOR | 4014 | 207720_at | -0.1129 | 0.2995 | |
| GSE26886 | LOR | 4014 | 207720_at | -0.2345 | 0.0856 | |
| GSE29001 | LOR | 4014 | 207720_at | 0.3435 | 0.5914 | |
| GSE38129 | LOR | 4014 | 207720_at | 0.3278 | 0.4815 | |
| GSE45670 | LOR | 4014 | 207720_at | 0.6757 | 0.4035 | |
| GSE53622 | LOR | 4014 | 35664 | 0.0020 | 0.9964 | |
| GSE53624 | LOR | 4014 | 35664 | -0.4871 | 0.1735 | |
| GSE63941 | LOR | 4014 | 207720_at | 0.1706 | 0.2775 | |
| GSE77861 | LOR | 4014 | 207720_at | -0.2342 | 0.1209 | |
| GSE97050 | LOR | 4014 | A_23_P34452 | 1.3154 | 0.3143 | |
| TCGA | LOR | 4014 | RNAseq | 1.3635 | 0.0471 |
Upregulated datasets: 1; Downregulated datasets: 0.
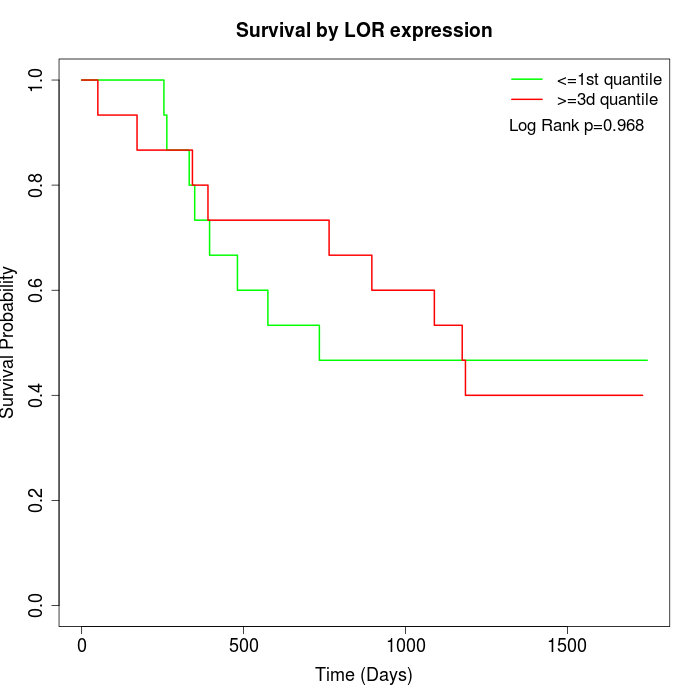
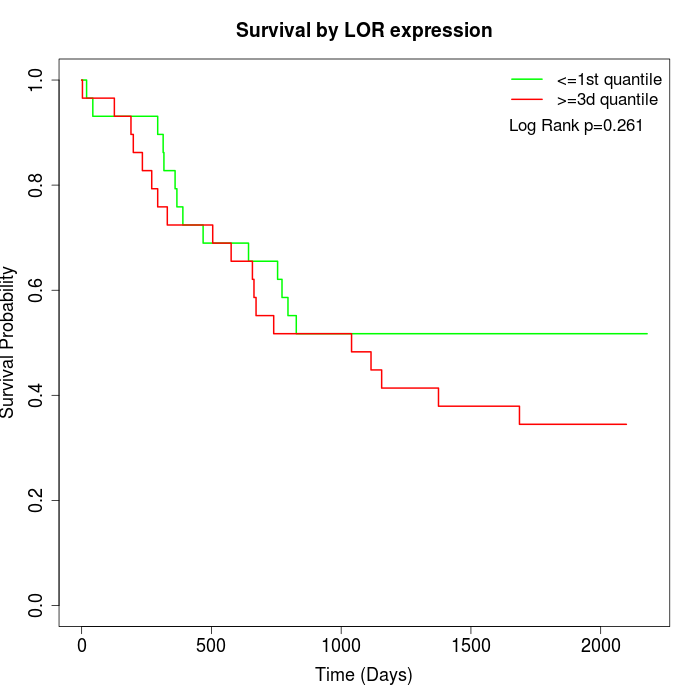
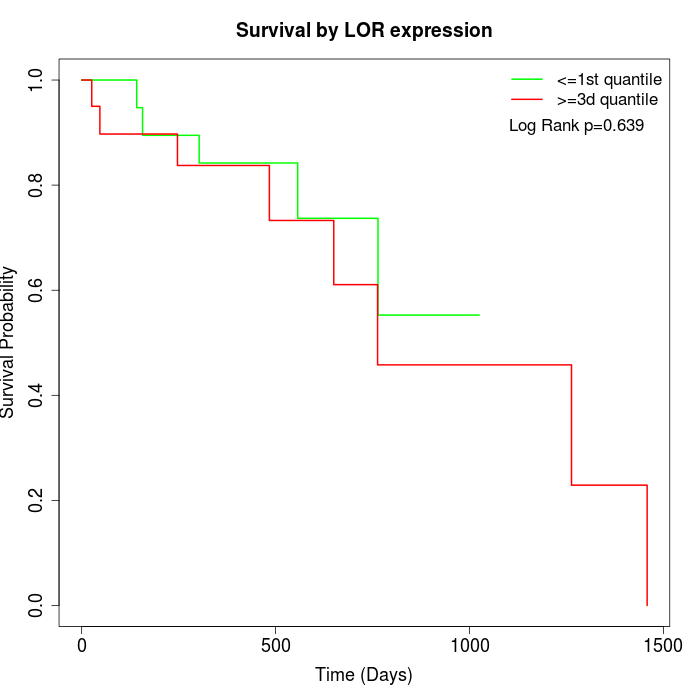
Survival by LOR expression:
Note: Click image to view full size file.
Copy number change of LOR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LOR | 4014 | 14 | 0 | 16 | |
| GSE20123 | LOR | 4014 | 14 | 0 | 16 | |
| GSE43470 | LOR | 4014 | 6 | 2 | 35 | |
| GSE46452 | LOR | 4014 | 2 | 1 | 56 | |
| GSE47630 | LOR | 4014 | 15 | 0 | 25 | |
| GSE54993 | LOR | 4014 | 0 | 4 | 66 | |
| GSE54994 | LOR | 4014 | 16 | 0 | 37 | |
| GSE60625 | LOR | 4014 | 0 | 0 | 11 | |
| GSE74703 | LOR | 4014 | 6 | 1 | 29 | |
| GSE74704 | LOR | 4014 | 7 | 0 | 13 | |
| TCGA | LOR | 4014 | 39 | 2 | 55 |
Total number of gains: 119; Total number of losses: 10; Total Number of normals: 359.
Somatic mutations of LOR:
Generating mutation plots.
Highly correlated genes for LOR:
Showing top 20/46 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LOR | CTCFL | 0.789502 | 3 | 0 | 3 |
| LOR | CSNK2A1 | 0.692707 | 4 | 0 | 3 |
| LOR | CEBPA | 0.689572 | 3 | 0 | 3 |
| LOR | SCN8A | 0.670872 | 3 | 0 | 3 |
| LOR | CCDC120 | 0.663399 | 3 | 0 | 3 |
| LOR | ASPRV1 | 0.660182 | 5 | 0 | 3 |
| LOR | BRDT | 0.633566 | 4 | 0 | 3 |
| LOR | CALB2 | 0.612377 | 4 | 0 | 3 |
| LOR | WNT11 | 0.603578 | 4 | 0 | 3 |
| LOR | TYSND1 | 0.603348 | 3 | 0 | 3 |
| LOR | FLG2 | 0.603319 | 6 | 0 | 5 |
| LOR | LCE2B | 0.589609 | 6 | 0 | 4 |
| LOR | TCEANC2 | 0.587844 | 5 | 0 | 3 |
| LOR | LRBA | 0.584131 | 3 | 0 | 3 |
| LOR | SAMD10 | 0.574629 | 3 | 0 | 3 |
| LOR | PPP1R3D | 0.571161 | 4 | 0 | 3 |
| LOR | SYPL1 | 0.566751 | 3 | 0 | 3 |
| LOR | SHB | 0.565977 | 4 | 0 | 3 |
| LOR | HOXC8 | 0.565793 | 4 | 0 | 3 |
| LOR | HSD17B10 | 0.564629 | 4 | 0 | 3 |
For details and further investigation, click here