| Full name: catenin delta 2 | Alias Symbol: NPRAP|GT24 | ||
| Type: protein-coding gene | Cytoband: 5p15.2 | ||
| Entrez ID: 1501 | HGNC ID: HGNC:2516 | Ensembl Gene: ENSG00000169862 | OMIM ID: 604275 |
| Drug and gene relationship at DGIdb | |||
Expression of CTNND2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CTNND2 | 1501 | 209617_s_at | -0.0703 | 0.7808 | |
| GSE20347 | CTNND2 | 1501 | 209617_s_at | 0.0266 | 0.6624 | |
| GSE23400 | CTNND2 | 1501 | 209617_s_at | -0.0741 | 0.0000 | |
| GSE26886 | CTNND2 | 1501 | 1566440_at | -0.1171 | 0.3590 | |
| GSE29001 | CTNND2 | 1501 | 209617_s_at | -0.0115 | 0.9484 | |
| GSE38129 | CTNND2 | 1501 | 209617_s_at | -0.0826 | 0.1470 | |
| GSE45670 | CTNND2 | 1501 | 209617_s_at | -0.1349 | 0.0852 | |
| GSE53622 | CTNND2 | 1501 | 80838 | -0.2934 | 0.0072 | |
| GSE53624 | CTNND2 | 1501 | 80838 | -0.2164 | 0.0398 | |
| GSE63941 | CTNND2 | 1501 | 209617_s_at | -0.1105 | 0.7651 | |
| GSE77861 | CTNND2 | 1501 | 1566440_at | -0.0247 | 0.8076 | |
| SRP133303 | CTNND2 | 1501 | RNAseq | -2.2308 | 0.0001 | |
| TCGA | CTNND2 | 1501 | RNAseq | -2.7998 | 0.0006 |
Upregulated datasets: 0; Downregulated datasets: 2.
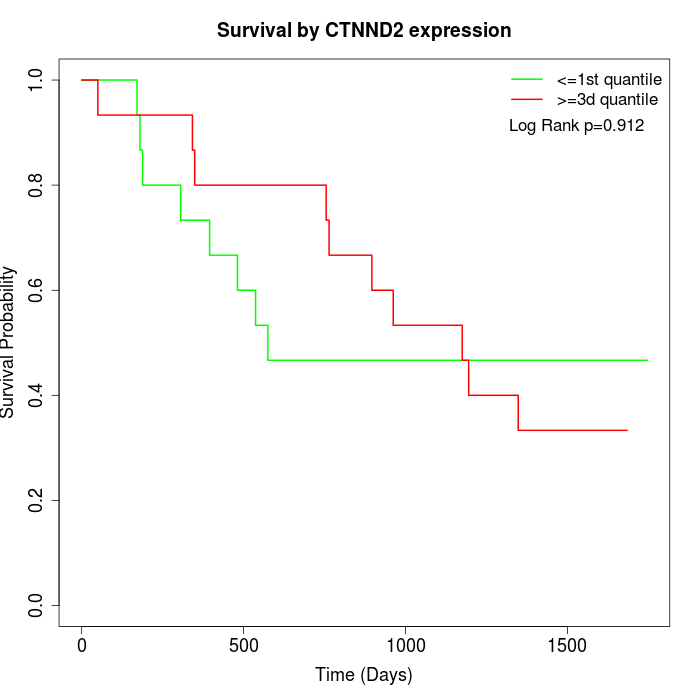
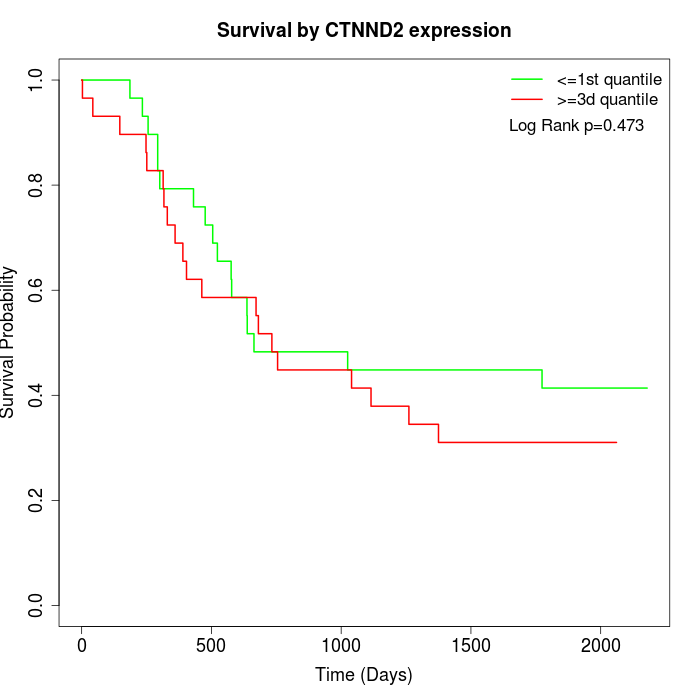
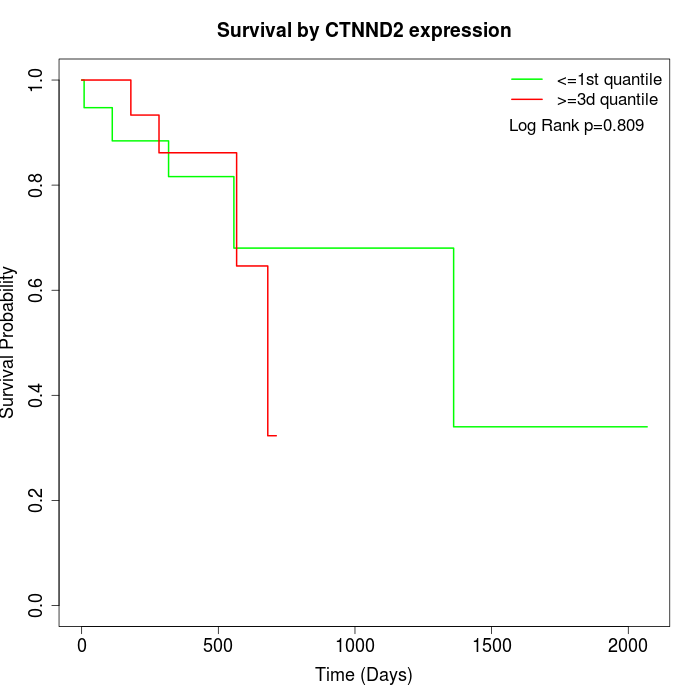
Survival by CTNND2 expression:
Note: Click image to view full size file.
Copy number change of CTNND2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CTNND2 | 1501 | 12 | 0 | 18 | |
| GSE20123 | CTNND2 | 1501 | 12 | 0 | 18 | |
| GSE43470 | CTNND2 | 1501 | 18 | 0 | 25 | |
| GSE46452 | CTNND2 | 1501 | 6 | 22 | 31 | |
| GSE47630 | CTNND2 | 1501 | 6 | 11 | 23 | |
| GSE54993 | CTNND2 | 1501 | 5 | 5 | 60 | |
| GSE54994 | CTNND2 | 1501 | 29 | 1 | 23 | |
| GSE60625 | CTNND2 | 1501 | 0 | 0 | 11 | |
| GSE74703 | CTNND2 | 1501 | 14 | 0 | 22 | |
| GSE74704 | CTNND2 | 1501 | 11 | 0 | 9 | |
| TCGA | CTNND2 | 1501 | 58 | 3 | 35 |
Total number of gains: 171; Total number of losses: 42; Total Number of normals: 275.
Somatic mutations of CTNND2:
Generating mutation plots.
Highly correlated genes for CTNND2:
Showing top 20/119 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CTNND2 | MAPRE3 | 0.689064 | 3 | 0 | 3 |
| CTNND2 | PDZD3 | 0.661661 | 4 | 0 | 4 |
| CTNND2 | MCF2 | 0.633874 | 3 | 0 | 3 |
| CTNND2 | KLRG1 | 0.621512 | 3 | 0 | 3 |
| CTNND2 | KCNMB2 | 0.616263 | 4 | 0 | 3 |
| CTNND2 | ECEL1 | 0.612596 | 4 | 0 | 4 |
| CTNND2 | ESRRB | 0.612428 | 3 | 0 | 3 |
| CTNND2 | KLC4 | 0.606136 | 3 | 0 | 3 |
| CTNND2 | RXRG | 0.604367 | 4 | 0 | 4 |
| CTNND2 | CHD5 | 0.603093 | 4 | 0 | 3 |
| CTNND2 | COL4A3 | 0.602938 | 5 | 0 | 4 |
| CTNND2 | IL17RC | 0.598537 | 3 | 0 | 3 |
| CTNND2 | SYP | 0.595117 | 4 | 0 | 3 |
| CTNND2 | CPS1-IT1 | 0.587566 | 5 | 0 | 3 |
| CTNND2 | CSN3 | 0.586209 | 4 | 0 | 3 |
| CTNND2 | LCT | 0.582273 | 3 | 0 | 3 |
| CTNND2 | SSC5D | 0.580989 | 3 | 0 | 3 |
| CTNND2 | PTH | 0.580207 | 4 | 0 | 3 |
| CTNND2 | ZNF582-AS1 | 0.579185 | 4 | 0 | 3 |
| CTNND2 | SLC22A2 | 0.576541 | 3 | 0 | 3 |
For details and further investigation, click here