| Full name: lipase maturation factor 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 22q13.33 | ||
| Entrez ID: 91289 | HGNC ID: HGNC:25096 | Ensembl Gene: ENSG00000100258 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LMF2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LMF2 | 91289 | 31837_at | -0.0081 | 0.9879 | |
| GSE20347 | LMF2 | 91289 | 31837_at | -0.0447 | 0.6403 | |
| GSE23400 | LMF2 | 91289 | 31837_at | 0.0443 | 0.2794 | |
| GSE26886 | LMF2 | 91289 | 31837_at | 0.1896 | 0.2976 | |
| GSE29001 | LMF2 | 91289 | 31837_at | -0.0246 | 0.9226 | |
| GSE38129 | LMF2 | 91289 | 31837_at | -0.0001 | 0.9990 | |
| GSE45670 | LMF2 | 91289 | 31837_at | 0.3003 | 0.0000 | |
| GSE53622 | LMF2 | 91289 | 50224 | 0.2695 | 0.0000 | |
| GSE53624 | LMF2 | 91289 | 50224 | 0.3042 | 0.0000 | |
| GSE63941 | LMF2 | 91289 | 31837_at | 0.0424 | 0.9096 | |
| GSE77861 | LMF2 | 91289 | 31837_at | 0.3034 | 0.0569 | |
| GSE97050 | LMF2 | 91289 | A_33_P3367642 | 0.3294 | 0.2151 | |
| SRP007169 | LMF2 | 91289 | RNAseq | 0.1332 | 0.7615 | |
| SRP008496 | LMF2 | 91289 | RNAseq | 0.0003 | 0.9988 | |
| SRP064894 | LMF2 | 91289 | RNAseq | 1.0288 | 0.0000 | |
| SRP133303 | LMF2 | 91289 | RNAseq | 0.2814 | 0.1338 | |
| SRP159526 | LMF2 | 91289 | RNAseq | 0.3353 | 0.0753 | |
| SRP193095 | LMF2 | 91289 | RNAseq | 0.4535 | 0.0001 | |
| SRP219564 | LMF2 | 91289 | RNAseq | 0.4796 | 0.1389 | |
| TCGA | LMF2 | 91289 | RNAseq | 0.1324 | 0.0100 |
Upregulated datasets: 1; Downregulated datasets: 0.
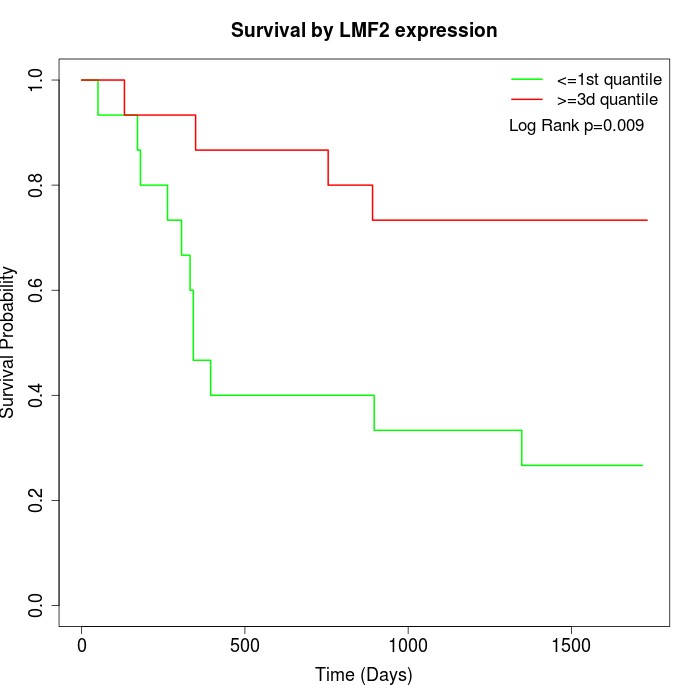
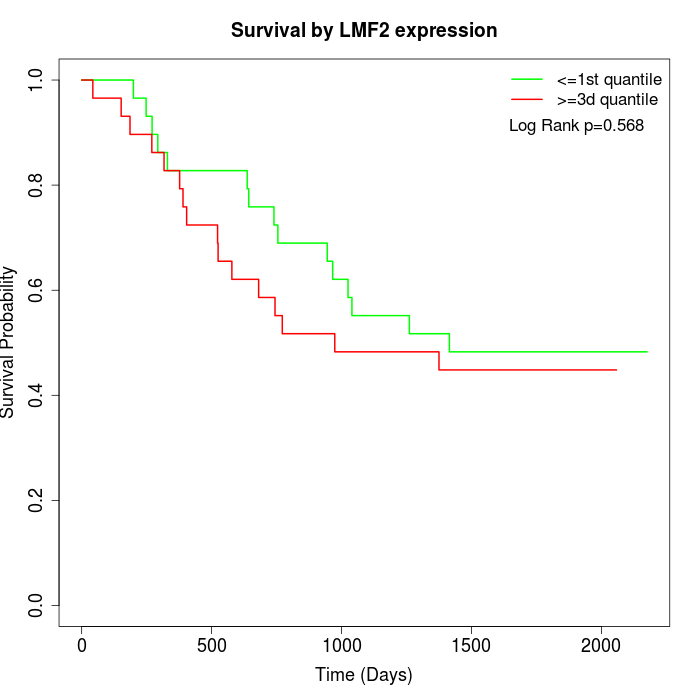
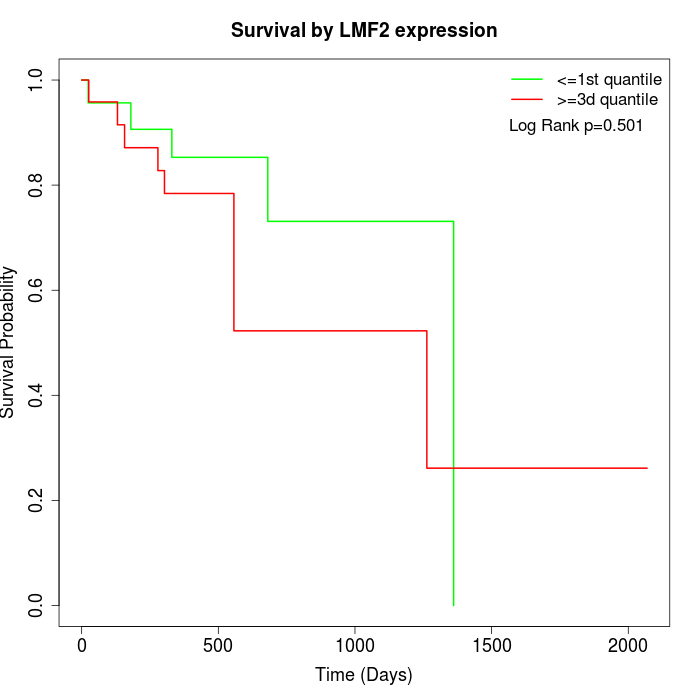
Survival by LMF2 expression:
Note: Click image to view full size file.
Copy number change of LMF2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LMF2 | 91289 | 3 | 8 | 19 | |
| GSE20123 | LMF2 | 91289 | 3 | 8 | 19 | |
| GSE43470 | LMF2 | 91289 | 2 | 8 | 33 | |
| GSE46452 | LMF2 | 91289 | 31 | 2 | 26 | |
| GSE47630 | LMF2 | 91289 | 9 | 4 | 27 | |
| GSE54993 | LMF2 | 91289 | 4 | 6 | 60 | |
| GSE54994 | LMF2 | 91289 | 11 | 8 | 34 | |
| GSE60625 | LMF2 | 91289 | 5 | 0 | 6 | |
| GSE74703 | LMF2 | 91289 | 2 | 6 | 28 | |
| GSE74704 | LMF2 | 91289 | 1 | 4 | 15 | |
| TCGA | LMF2 | 91289 | 26 | 16 | 54 |
Total number of gains: 97; Total number of losses: 70; Total Number of normals: 321.
Somatic mutations of LMF2:
Generating mutation plots.
Highly correlated genes for LMF2:
Showing top 20/523 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LMF2 | SNAPC2 | 0.757824 | 3 | 0 | 3 |
| LMF2 | LSM12 | 0.73762 | 3 | 0 | 3 |
| LMF2 | ZNF747 | 0.729143 | 3 | 0 | 3 |
| LMF2 | RAB2B | 0.726385 | 3 | 0 | 3 |
| LMF2 | PFDN1 | 0.725773 | 3 | 0 | 3 |
| LMF2 | PPP6R1 | 0.72015 | 3 | 0 | 3 |
| LMF2 | GRIN3B | 0.718494 | 3 | 0 | 3 |
| LMF2 | TERF2 | 0.716718 | 3 | 0 | 3 |
| LMF2 | ZNF827 | 0.710229 | 3 | 0 | 3 |
| LMF2 | FRMD8 | 0.703116 | 3 | 0 | 3 |
| LMF2 | FAM43A | 0.69477 | 3 | 0 | 3 |
| LMF2 | DENND3 | 0.694327 | 3 | 0 | 3 |
| LMF2 | PARP9 | 0.694003 | 3 | 0 | 3 |
| LMF2 | YLPM1 | 0.691667 | 3 | 0 | 3 |
| LMF2 | PUF60 | 0.691481 | 3 | 0 | 3 |
| LMF2 | AAK1 | 0.690517 | 5 | 0 | 5 |
| LMF2 | FUS | 0.690476 | 3 | 0 | 3 |
| LMF2 | ALOXE3 | 0.688728 | 3 | 0 | 3 |
| LMF2 | INTS1 | 0.687808 | 4 | 0 | 4 |
| LMF2 | C1orf68 | 0.686366 | 3 | 0 | 3 |
For details and further investigation, click here