| Full name: cullin 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7q36.1 | ||
| Entrez ID: 8454 | HGNC ID: HGNC:2551 | Ensembl Gene: ENSG00000055130 | OMIM ID: 603134 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CUL1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle | |
| hsa04114 | Oocyte meiosis | |
| hsa04340 | Hedgehog signaling pathway | |
| hsa04350 | TGF-beta signaling pathway | |
| hsa04710 | Circadian rhythm |
Expression of CUL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CUL1 | 8454 | 207614_s_at | -0.0294 | 0.9548 | |
| GSE20347 | CUL1 | 8454 | 207614_s_at | 0.2469 | 0.0910 | |
| GSE23400 | CUL1 | 8454 | 207614_s_at | 0.2250 | 0.0013 | |
| GSE26886 | CUL1 | 8454 | 207614_s_at | -0.5372 | 0.0617 | |
| GSE29001 | CUL1 | 8454 | 207614_s_at | 0.2480 | 0.6252 | |
| GSE38129 | CUL1 | 8454 | 207614_s_at | 0.2198 | 0.0481 | |
| GSE45670 | CUL1 | 8454 | 207614_s_at | 0.0269 | 0.8802 | |
| GSE53622 | CUL1 | 8454 | 55468 | 0.2961 | 0.0000 | |
| GSE53624 | CUL1 | 8454 | 55468 | 0.3473 | 0.0000 | |
| GSE63941 | CUL1 | 8454 | 207614_s_at | -0.1916 | 0.5376 | |
| GSE77861 | CUL1 | 8454 | 207614_s_at | 0.5595 | 0.0021 | |
| GSE97050 | CUL1 | 8454 | A_24_P303589 | -0.0085 | 0.9811 | |
| SRP007169 | CUL1 | 8454 | RNAseq | 0.8404 | 0.0164 | |
| SRP008496 | CUL1 | 8454 | RNAseq | 0.6549 | 0.0020 | |
| SRP064894 | CUL1 | 8454 | RNAseq | 0.2752 | 0.0274 | |
| SRP133303 | CUL1 | 8454 | RNAseq | 0.2864 | 0.0202 | |
| SRP159526 | CUL1 | 8454 | RNAseq | 0.0911 | 0.7189 | |
| SRP193095 | CUL1 | 8454 | RNAseq | 0.0195 | 0.8218 | |
| SRP219564 | CUL1 | 8454 | RNAseq | -0.3598 | 0.2213 | |
| TCGA | CUL1 | 8454 | RNAseq | 0.0107 | 0.8169 |
Upregulated datasets: 0; Downregulated datasets: 0.
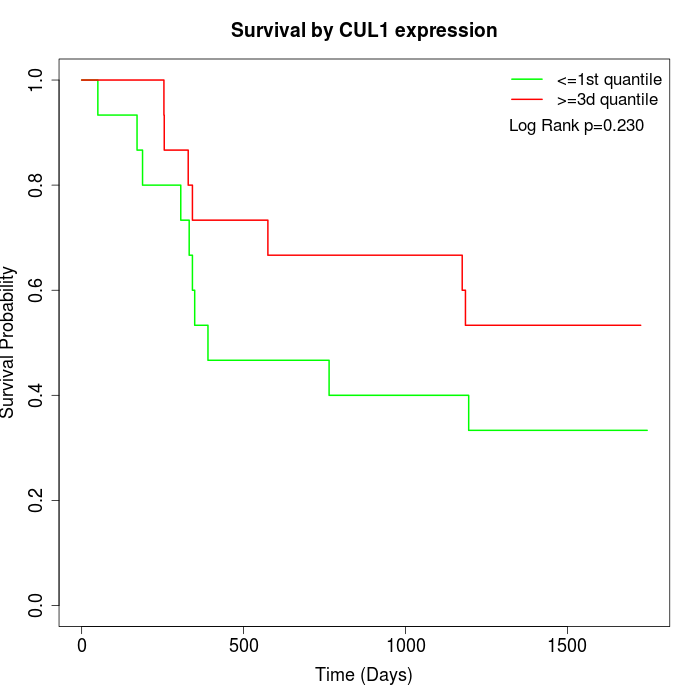
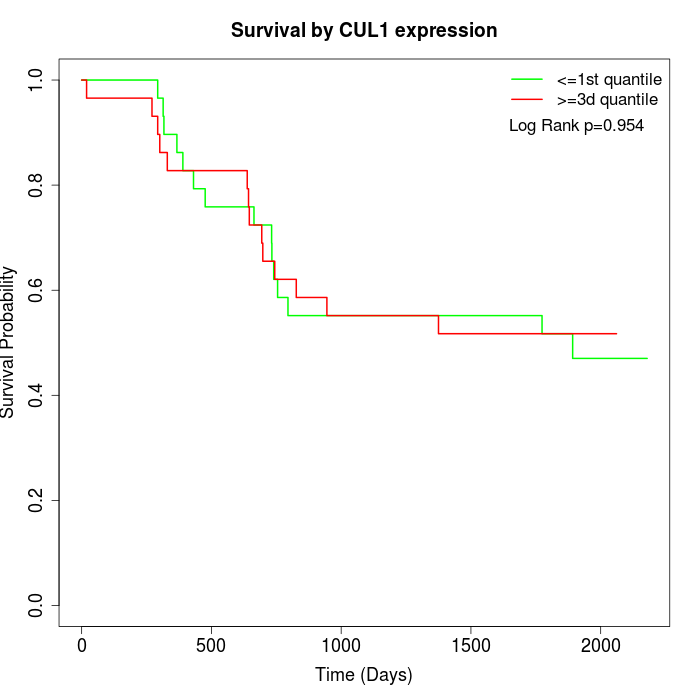
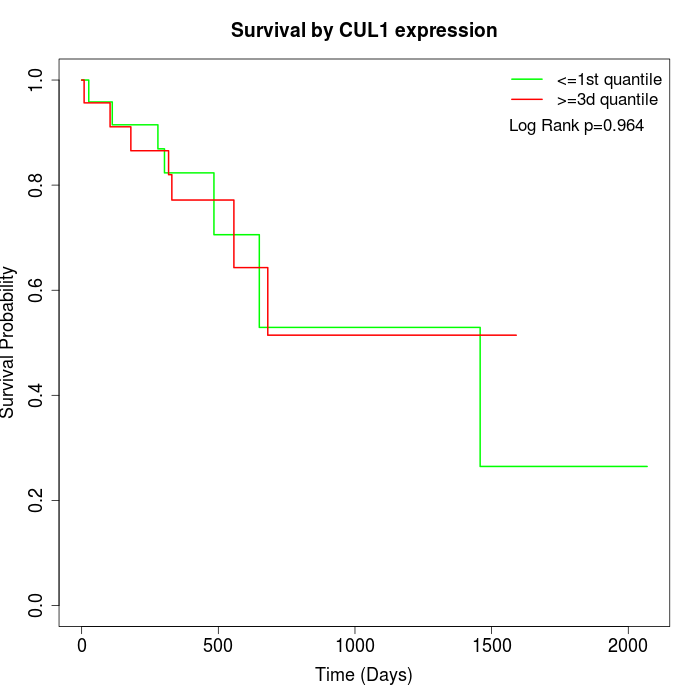
Survival by CUL1 expression:
Note: Click image to view full size file.
Copy number change of CUL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CUL1 | 8454 | 2 | 3 | 25 | |
| GSE20123 | CUL1 | 8454 | 2 | 3 | 25 | |
| GSE43470 | CUL1 | 8454 | 2 | 5 | 36 | |
| GSE46452 | CUL1 | 8454 | 7 | 2 | 50 | |
| GSE47630 | CUL1 | 8454 | 6 | 8 | 26 | |
| GSE54993 | CUL1 | 8454 | 3 | 4 | 63 | |
| GSE54994 | CUL1 | 8454 | 5 | 9 | 39 | |
| GSE60625 | CUL1 | 8454 | 0 | 0 | 11 | |
| GSE74703 | CUL1 | 8454 | 2 | 4 | 30 | |
| GSE74704 | CUL1 | 8454 | 1 | 3 | 16 | |
| TCGA | CUL1 | 8454 | 25 | 27 | 44 |
Total number of gains: 55; Total number of losses: 68; Total Number of normals: 365.
Somatic mutations of CUL1:
Generating mutation plots.
Highly correlated genes for CUL1:
Showing top 20/784 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CUL1 | DLG5 | 0.80954 | 3 | 0 | 3 |
| CUL1 | ATL1 | 0.79357 | 3 | 0 | 3 |
| CUL1 | GOLIM4 | 0.76754 | 3 | 0 | 3 |
| CUL1 | SENP8 | 0.764904 | 3 | 0 | 3 |
| CUL1 | UFC1 | 0.760442 | 3 | 0 | 3 |
| CUL1 | SLC30A5 | 0.755718 | 4 | 0 | 3 |
| CUL1 | THAP5 | 0.755315 | 4 | 0 | 4 |
| CUL1 | SDHD | 0.749457 | 3 | 0 | 3 |
| CUL1 | UBE2N | 0.748541 | 3 | 0 | 3 |
| CUL1 | HEATR5A | 0.744443 | 3 | 0 | 3 |
| CUL1 | SPPL2A | 0.742137 | 3 | 0 | 3 |
| CUL1 | C6orf89 | 0.736933 | 3 | 0 | 3 |
| CUL1 | TSEN2 | 0.72761 | 3 | 0 | 3 |
| CUL1 | SERINC1 | 0.72248 | 3 | 0 | 3 |
| CUL1 | TMEM205 | 0.722039 | 4 | 0 | 4 |
| CUL1 | RHEB | 0.721792 | 13 | 0 | 13 |
| CUL1 | KLHL20 | 0.719462 | 3 | 0 | 3 |
| CUL1 | ZYG11B | 0.718862 | 3 | 0 | 3 |
| CUL1 | UNC45A | 0.717629 | 3 | 0 | 3 |
| CUL1 | GPHN | 0.71751 | 3 | 0 | 3 |
For details and further investigation, click here