| Full name: cullin 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10p11.21 | ||
| Entrez ID: 8453 | HGNC ID: HGNC:2552 | Ensembl Gene: ENSG00000108094 | OMIM ID: 603135 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CUL2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05200 | Pathways in cancer |
Expression of CUL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CUL2 | 8453 | 203079_s_at | -0.0720 | 0.8647 | |
| GSE20347 | CUL2 | 8453 | 203079_s_at | 0.1255 | 0.4739 | |
| GSE23400 | CUL2 | 8453 | 203079_s_at | 0.2684 | 0.0000 | |
| GSE26886 | CUL2 | 8453 | 203079_s_at | -0.5599 | 0.0065 | |
| GSE29001 | CUL2 | 8453 | 203079_s_at | 0.1591 | 0.6828 | |
| GSE38129 | CUL2 | 8453 | 203079_s_at | 0.1317 | 0.3514 | |
| GSE45670 | CUL2 | 8453 | 203079_s_at | 0.2045 | 0.3694 | |
| GSE53622 | CUL2 | 8453 | 150081 | 0.1092 | 0.2514 | |
| GSE53624 | CUL2 | 8453 | 150081 | 0.1373 | 0.0902 | |
| GSE63941 | CUL2 | 8453 | 203079_s_at | -0.5500 | 0.3294 | |
| GSE77861 | CUL2 | 8453 | 203079_s_at | -0.0847 | 0.7596 | |
| GSE97050 | CUL2 | 8453 | A_24_P252794 | 0.1307 | 0.5340 | |
| SRP007169 | CUL2 | 8453 | RNAseq | 0.0218 | 0.9546 | |
| SRP008496 | CUL2 | 8453 | RNAseq | 0.4640 | 0.1776 | |
| SRP064894 | CUL2 | 8453 | RNAseq | 0.0080 | 0.9585 | |
| SRP133303 | CUL2 | 8453 | RNAseq | 0.1412 | 0.2168 | |
| SRP159526 | CUL2 | 8453 | RNAseq | -0.1664 | 0.5486 | |
| SRP193095 | CUL2 | 8453 | RNAseq | -0.1279 | 0.1055 | |
| SRP219564 | CUL2 | 8453 | RNAseq | -0.4434 | 0.1417 | |
| TCGA | CUL2 | 8453 | RNAseq | 0.0424 | 0.4682 |
Upregulated datasets: 0; Downregulated datasets: 0.
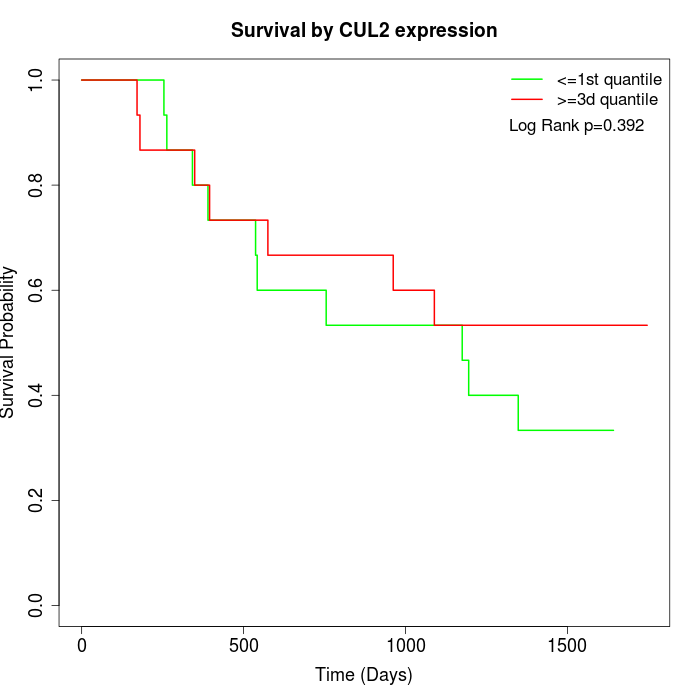
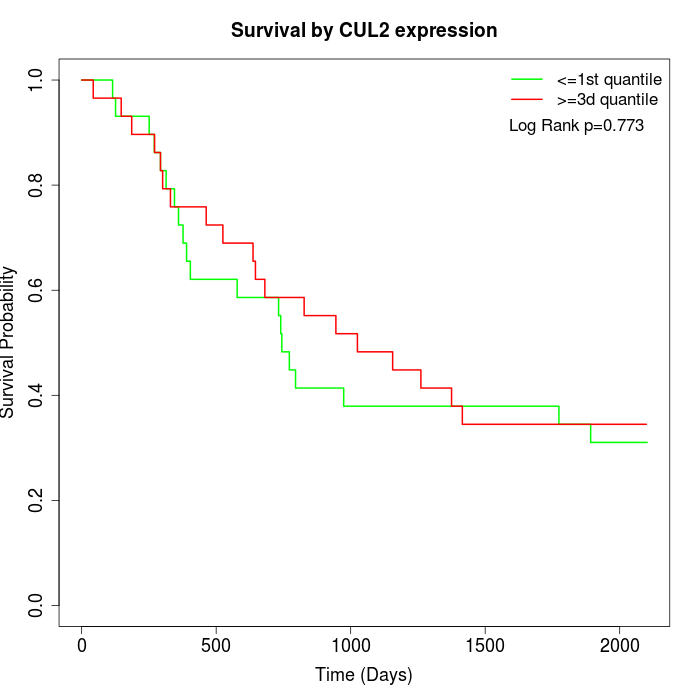
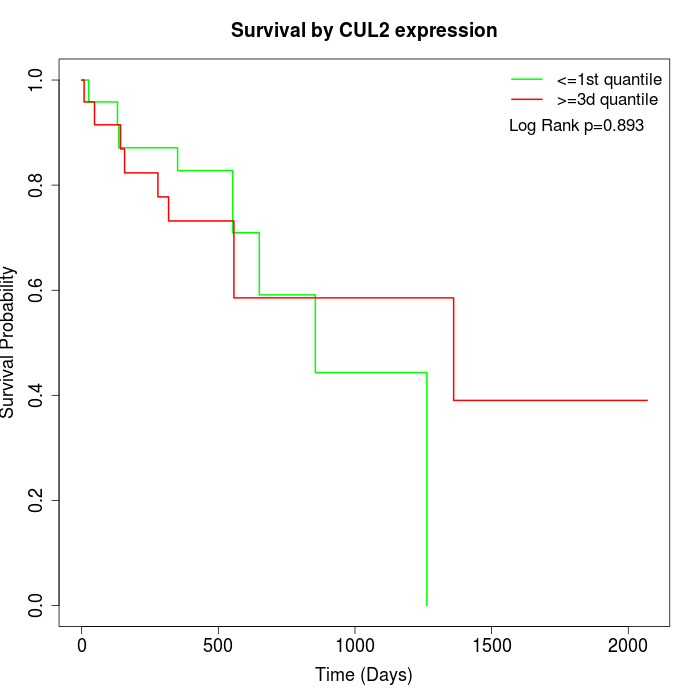
Survival by CUL2 expression:
Note: Click image to view full size file.
Copy number change of CUL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CUL2 | 8453 | 5 | 7 | 18 | |
| GSE20123 | CUL2 | 8453 | 5 | 6 | 19 | |
| GSE43470 | CUL2 | 8453 | 3 | 3 | 37 | |
| GSE46452 | CUL2 | 8453 | 2 | 13 | 44 | |
| GSE47630 | CUL2 | 8453 | 7 | 10 | 23 | |
| GSE54993 | CUL2 | 8453 | 8 | 1 | 61 | |
| GSE54994 | CUL2 | 8453 | 2 | 10 | 41 | |
| GSE60625 | CUL2 | 8453 | 0 | 0 | 11 | |
| GSE74703 | CUL2 | 8453 | 2 | 1 | 33 | |
| GSE74704 | CUL2 | 8453 | 0 | 5 | 15 | |
| TCGA | CUL2 | 8453 | 19 | 24 | 53 |
Total number of gains: 53; Total number of losses: 80; Total Number of normals: 355.
Somatic mutations of CUL2:
Generating mutation plots.
Highly correlated genes for CUL2:
Showing top 20/598 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CUL2 | CNOT4 | 0.786048 | 3 | 0 | 3 |
| CUL2 | PRPF38B | 0.78592 | 3 | 0 | 3 |
| CUL2 | AP3M1 | 0.769714 | 3 | 0 | 3 |
| CUL2 | WDR20 | 0.76852 | 3 | 0 | 3 |
| CUL2 | LGALSL | 0.768515 | 3 | 0 | 3 |
| CUL2 | LANCL1 | 0.767827 | 4 | 0 | 4 |
| CUL2 | TMEM41B | 0.767742 | 3 | 0 | 3 |
| CUL2 | NR2C2 | 0.758243 | 4 | 0 | 3 |
| CUL2 | TYW3 | 0.756788 | 3 | 0 | 3 |
| CUL2 | SARNP | 0.756668 | 3 | 0 | 3 |
| CUL2 | ATG4C | 0.75602 | 3 | 0 | 3 |
| CUL2 | EIF2B2 | 0.754507 | 3 | 0 | 3 |
| CUL2 | SNX18 | 0.752244 | 3 | 0 | 3 |
| CUL2 | MCMBP | 0.741269 | 3 | 0 | 3 |
| CUL2 | SLC30A9 | 0.740337 | 3 | 0 | 3 |
| CUL2 | FAHD1 | 0.737272 | 3 | 0 | 3 |
| CUL2 | RRN3 | 0.733768 | 4 | 0 | 4 |
| CUL2 | GMCL1 | 0.73181 | 3 | 0 | 3 |
| CUL2 | TMEM218 | 0.72837 | 3 | 0 | 3 |
| CUL2 | PPP2CA | 0.725775 | 3 | 0 | 3 |
For details and further investigation, click here