| Full name: cytochrome P450 family 4 subfamily F member 12 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 19p13.12 | ||
| Entrez ID: 66002 | HGNC ID: HGNC:18857 | Ensembl Gene: ENSG00000186204 | OMIM ID: 611485 |
| Drug and gene relationship at DGIdb | |||
Expression of CYP4F12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CYP4F12 | 66002 | 206539_s_at | -1.1651 | 0.0088 | |
| GSE20347 | CYP4F12 | 66002 | 206539_s_at | -0.8889 | 0.0000 | |
| GSE23400 | CYP4F12 | 66002 | 206539_s_at | -0.8514 | 0.0000 | |
| GSE26886 | CYP4F12 | 66002 | 206539_s_at | -1.0940 | 0.0000 | |
| GSE29001 | CYP4F12 | 66002 | 206539_s_at | -1.6096 | 0.0001 | |
| GSE38129 | CYP4F12 | 66002 | 206539_s_at | -1.0581 | 0.0014 | |
| GSE45670 | CYP4F12 | 66002 | 206539_s_at | -0.8708 | 0.0047 | |
| GSE53622 | CYP4F12 | 66002 | 6472 | -0.4858 | 0.1324 | |
| GSE53624 | CYP4F12 | 66002 | 6472 | -1.1281 | 0.0000 | |
| GSE63941 | CYP4F12 | 66002 | 206539_s_at | 1.6195 | 0.0002 | |
| GSE77861 | CYP4F12 | 66002 | 206539_s_at | -0.6468 | 0.0149 | |
| GSE97050 | CYP4F12 | 66002 | A_23_P108280 | -0.4822 | 0.2289 | |
| SRP007169 | CYP4F12 | 66002 | RNAseq | -3.8559 | 0.0000 | |
| SRP008496 | CYP4F12 | 66002 | RNAseq | -3.5957 | 0.0000 | |
| SRP064894 | CYP4F12 | 66002 | RNAseq | -2.7208 | 0.0000 | |
| SRP133303 | CYP4F12 | 66002 | RNAseq | -3.7279 | 0.0000 | |
| SRP159526 | CYP4F12 | 66002 | RNAseq | -3.5105 | 0.0000 | |
| SRP193095 | CYP4F12 | 66002 | RNAseq | -2.3851 | 0.0000 | |
| SRP219564 | CYP4F12 | 66002 | RNAseq | -3.3568 | 0.0006 | |
| TCGA | CYP4F12 | 66002 | RNAseq | -1.1503 | 0.0178 |
Upregulated datasets: 1; Downregulated datasets: 13.
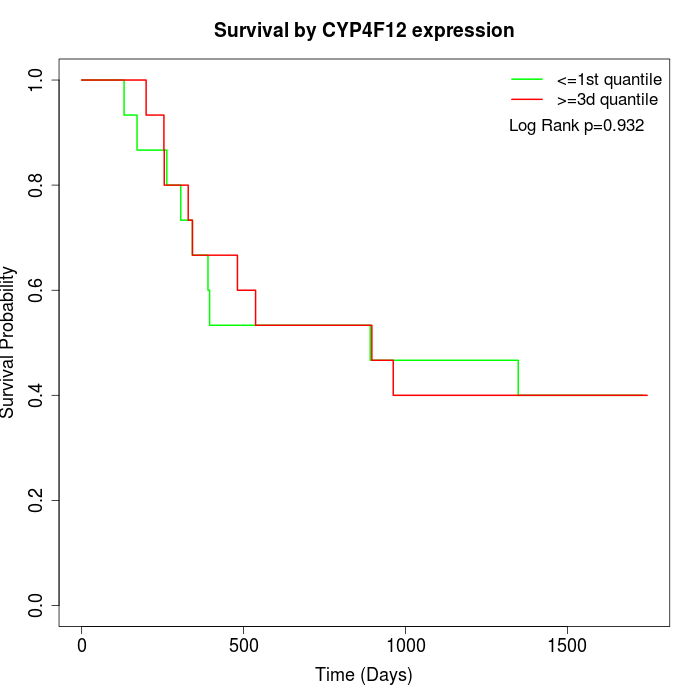
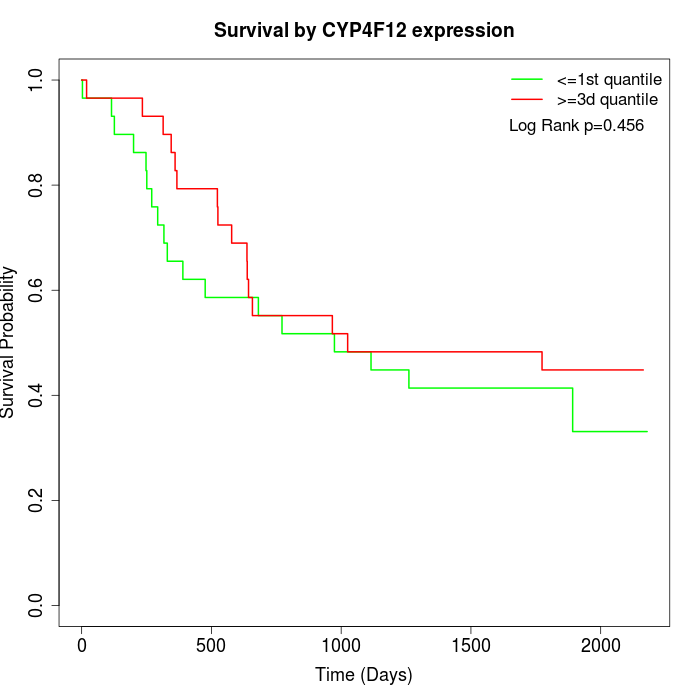
Survival by CYP4F12 expression:
Note: Click image to view full size file.
Copy number change of CYP4F12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CYP4F12 | 66002 | 4 | 2 | 24 | |
| GSE20123 | CYP4F12 | 66002 | 3 | 1 | 26 | |
| GSE43470 | CYP4F12 | 66002 | 2 | 5 | 36 | |
| GSE46452 | CYP4F12 | 66002 | 47 | 1 | 11 | |
| GSE47630 | CYP4F12 | 66002 | 4 | 7 | 29 | |
| GSE54993 | CYP4F12 | 66002 | 15 | 4 | 51 | |
| GSE54994 | CYP4F12 | 66002 | 6 | 14 | 33 | |
| GSE60625 | CYP4F12 | 66002 | 9 | 0 | 2 | |
| GSE74703 | CYP4F12 | 66002 | 2 | 4 | 30 | |
| GSE74704 | CYP4F12 | 66002 | 0 | 1 | 19 | |
| TCGA | CYP4F12 | 66002 | 20 | 10 | 66 |
Total number of gains: 112; Total number of losses: 49; Total Number of normals: 327.
Somatic mutations of CYP4F12:
Generating mutation plots.
Highly correlated genes for CYP4F12:
Showing top 20/1093 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CYP4F12 | PADI1 | 0.768493 | 9 | 0 | 9 |
| CYP4F12 | FAM95B1 | 0.757125 | 5 | 0 | 5 |
| CYP4F12 | SFTA2 | 0.752803 | 4 | 0 | 4 |
| CYP4F12 | PAQR8 | 0.746458 | 4 | 0 | 4 |
| CYP4F12 | CES2 | 0.742197 | 9 | 0 | 9 |
| CYP4F12 | SNORA68 | 0.741298 | 5 | 0 | 4 |
| CYP4F12 | FUT6 | 0.737634 | 9 | 0 | 9 |
| CYP4F12 | FCHO2 | 0.73146 | 4 | 0 | 4 |
| CYP4F12 | CGNL1 | 0.727609 | 4 | 0 | 4 |
| CYP4F12 | SMPD2 | 0.71917 | 8 | 0 | 8 |
| CYP4F12 | SORT1 | 0.718506 | 8 | 0 | 8 |
| CYP4F12 | PRSS2 | 0.717662 | 9 | 0 | 9 |
| CYP4F12 | SPAG17 | 0.717598 | 5 | 0 | 5 |
| CYP4F12 | PPP1R7 | 0.715233 | 6 | 0 | 6 |
| CYP4F12 | CYP4F8 | 0.71222 | 5 | 0 | 4 |
| CYP4F12 | SAMD5 | 0.710901 | 6 | 0 | 5 |
| CYP4F12 | CNPPD1 | 0.710247 | 8 | 0 | 8 |
| CYP4F12 | SHROOM3 | 0.708923 | 4 | 0 | 4 |
| CYP4F12 | PLEKHA7 | 0.708664 | 5 | 0 | 5 |
| CYP4F12 | CRNN | 0.708101 | 10 | 0 | 8 |
For details and further investigation, click here