| Full name: DExD/H-box 60 like | Alias Symbol: FLJ31033 | ||
| Type: protein-coding gene | Cytoband: 4q32.3 | ||
| Entrez ID: 91351 | HGNC ID: HGNC:26429 | Ensembl Gene: ENSG00000181381 | OMIM ID: 616725 |
| Drug and gene relationship at DGIdb | |||
Expression of DDX60L:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DDX60L | 91351 | 228152_s_at | 0.7144 | 0.5107 | |
| GSE26886 | DDX60L | 91351 | 228152_s_at | 1.4215 | 0.0027 | |
| GSE45670 | DDX60L | 91351 | 228152_s_at | 1.6222 | 0.0007 | |
| GSE63941 | DDX60L | 91351 | 228152_s_at | -1.3811 | 0.2974 | |
| GSE77861 | DDX60L | 91351 | 228152_s_at | 0.5491 | 0.1187 | |
| GSE97050 | DDX60L | 91351 | A_23_P30069 | 0.9134 | 0.0835 | |
| SRP007169 | DDX60L | 91351 | RNAseq | 2.8698 | 0.0000 | |
| SRP008496 | DDX60L | 91351 | RNAseq | 2.2920 | 0.0000 | |
| SRP064894 | DDX60L | 91351 | RNAseq | 2.1691 | 0.0000 | |
| SRP133303 | DDX60L | 91351 | RNAseq | 1.5287 | 0.0000 | |
| SRP159526 | DDX60L | 91351 | RNAseq | -0.2543 | 0.6065 | |
| SRP193095 | DDX60L | 91351 | RNAseq | 0.6058 | 0.0850 | |
| SRP219564 | DDX60L | 91351 | RNAseq | 0.7213 | 0.2682 | |
| TCGA | DDX60L | 91351 | RNAseq | 0.0805 | 0.4808 |
Upregulated datasets: 6; Downregulated datasets: 0.
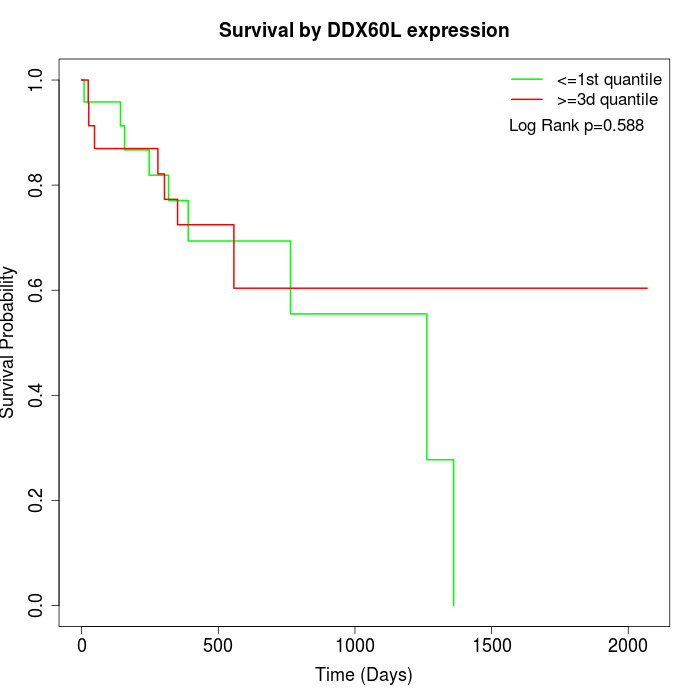
Survival by DDX60L expression:
Note: Click image to view full size file.
Copy number change of DDX60L:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DDX60L | 91351 | 1 | 11 | 18 | |
| GSE20123 | DDX60L | 91351 | 1 | 11 | 18 | |
| GSE43470 | DDX60L | 91351 | 0 | 13 | 30 | |
| GSE46452 | DDX60L | 91351 | 1 | 36 | 22 | |
| GSE47630 | DDX60L | 91351 | 0 | 22 | 18 | |
| GSE54993 | DDX60L | 91351 | 10 | 0 | 60 | |
| GSE54994 | DDX60L | 91351 | 1 | 13 | 39 | |
| GSE60625 | DDX60L | 91351 | 0 | 0 | 11 | |
| GSE74703 | DDX60L | 91351 | 0 | 11 | 25 | |
| GSE74704 | DDX60L | 91351 | 0 | 5 | 15 | |
| TCGA | DDX60L | 91351 | 12 | 33 | 51 |
Total number of gains: 26; Total number of losses: 155; Total Number of normals: 307.
Somatic mutations of DDX60L:
Generating mutation plots.
Highly correlated genes for DDX60L:
Showing top 20/661 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DDX60L | DDX60 | 0.836214 | 7 | 0 | 7 |
| DDX60L | SP110 | 0.787942 | 6 | 0 | 5 |
| DDX60L | STAT1 | 0.785708 | 7 | 0 | 6 |
| DDX60L | NMD3 | 0.785102 | 3 | 0 | 3 |
| DDX60L | LRCH3 | 0.784344 | 3 | 0 | 3 |
| DDX60L | UBTD2 | 0.780825 | 3 | 0 | 3 |
| DDX60L | IFI44 | 0.766297 | 7 | 0 | 7 |
| DDX60L | RPL26L1 | 0.765992 | 3 | 0 | 3 |
| DDX60L | GBP1 | 0.765612 | 6 | 0 | 6 |
| DDX60L | CD80 | 0.758376 | 3 | 0 | 3 |
| DDX60L | FAM126A | 0.753162 | 3 | 0 | 3 |
| DDX60L | HELZ2 | 0.75064 | 3 | 0 | 3 |
| DDX60L | SLC39A14 | 0.746278 | 3 | 0 | 3 |
| DDX60L | IFI6 | 0.740847 | 7 | 0 | 7 |
| DDX60L | AHR | 0.740024 | 3 | 0 | 3 |
| DDX60L | NEK3 | 0.738464 | 3 | 0 | 3 |
| DDX60L | RBM39 | 0.737803 | 3 | 0 | 3 |
| DDX60L | PLA2G7 | 0.735685 | 5 | 0 | 5 |
| DDX60L | CACYBP | 0.734155 | 3 | 0 | 3 |
| DDX60L | IFIH1 | 0.733963 | 7 | 0 | 6 |
For details and further investigation, click here