| Full name: DEAF1 transcription factor | Alias Symbol: NUDR|SPN|ZMYND5 | ||
| Type: protein-coding gene | Cytoband: 11p15.5 | ||
| Entrez ID: 10522 | HGNC ID: HGNC:14677 | Ensembl Gene: ENSG00000177030 | OMIM ID: 602635 |
| Drug and gene relationship at DGIdb | |||
Expression of DEAF1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DEAF1 | 10522 | 209407_s_at | -0.5331 | 0.1584 | |
| GSE20347 | DEAF1 | 10522 | 209407_s_at | -0.6778 | 0.0000 | |
| GSE23400 | DEAF1 | 10522 | 209407_s_at | -0.3913 | 0.0000 | |
| GSE26886 | DEAF1 | 10522 | 209407_s_at | -0.4492 | 0.0256 | |
| GSE29001 | DEAF1 | 10522 | 209407_s_at | -0.5120 | 0.0106 | |
| GSE38129 | DEAF1 | 10522 | 209407_s_at | -0.6160 | 0.0000 | |
| GSE45670 | DEAF1 | 10522 | 209407_s_at | -0.3156 | 0.0547 | |
| GSE53622 | DEAF1 | 10522 | 7075 | -0.6771 | 0.0000 | |
| GSE53624 | DEAF1 | 10522 | 7075 | -0.7722 | 0.0000 | |
| GSE63941 | DEAF1 | 10522 | 209407_s_at | -0.4711 | 0.3741 | |
| GSE77861 | DEAF1 | 10522 | 209407_s_at | -0.3042 | 0.3113 | |
| GSE97050 | DEAF1 | 10522 | A_23_P158533 | -0.0574 | 0.7921 | |
| SRP007169 | DEAF1 | 10522 | RNAseq | -2.6913 | 0.0000 | |
| SRP008496 | DEAF1 | 10522 | RNAseq | -2.2137 | 0.0000 | |
| SRP064894 | DEAF1 | 10522 | RNAseq | -0.7614 | 0.0008 | |
| SRP133303 | DEAF1 | 10522 | RNAseq | -1.0541 | 0.0000 | |
| SRP159526 | DEAF1 | 10522 | RNAseq | -1.1011 | 0.0000 | |
| SRP193095 | DEAF1 | 10522 | RNAseq | -1.0812 | 0.0000 | |
| SRP219564 | DEAF1 | 10522 | RNAseq | -1.0467 | 0.0163 | |
| TCGA | DEAF1 | 10522 | RNAseq | -0.2965 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 6.
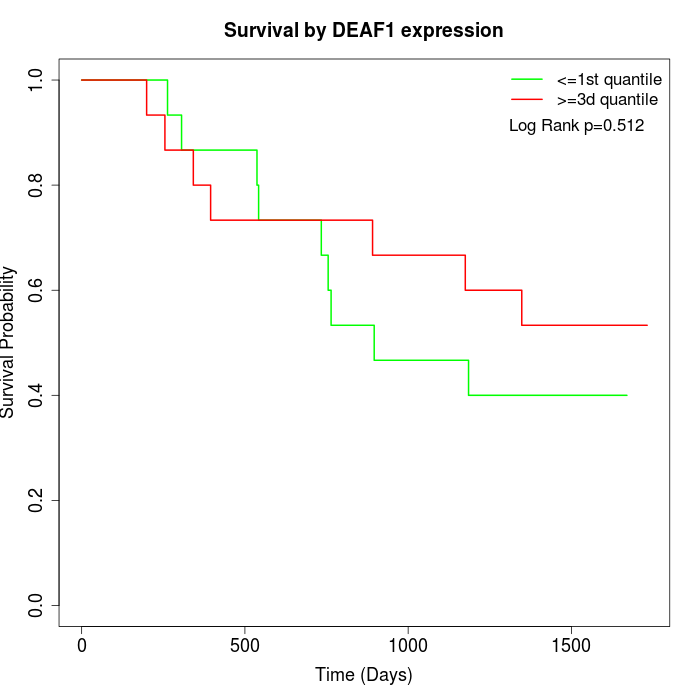
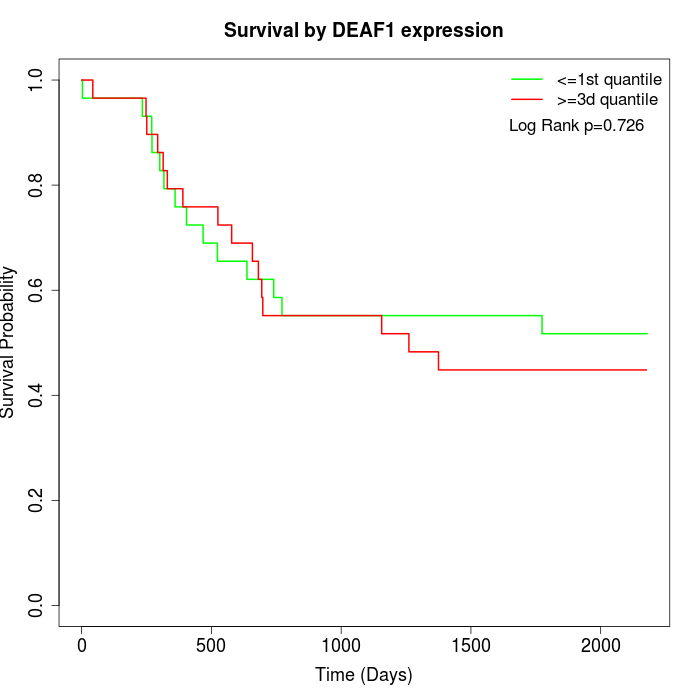
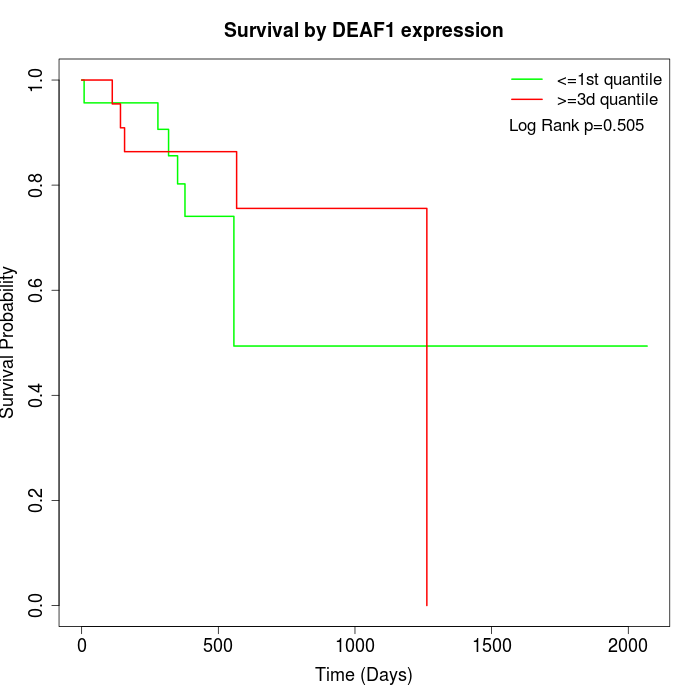
Survival by DEAF1 expression:
Note: Click image to view full size file.
Copy number change of DEAF1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DEAF1 | 10522 | 1 | 11 | 18 | |
| GSE20123 | DEAF1 | 10522 | 1 | 12 | 17 | |
| GSE43470 | DEAF1 | 10522 | 1 | 10 | 32 | |
| GSE46452 | DEAF1 | 10522 | 7 | 8 | 44 | |
| GSE47630 | DEAF1 | 10522 | 4 | 12 | 24 | |
| GSE54993 | DEAF1 | 10522 | 3 | 1 | 66 | |
| GSE54994 | DEAF1 | 10522 | 1 | 12 | 40 | |
| GSE60625 | DEAF1 | 10522 | 0 | 0 | 11 | |
| GSE74703 | DEAF1 | 10522 | 1 | 8 | 27 | |
| GSE74704 | DEAF1 | 10522 | 0 | 8 | 12 | |
| TCGA | DEAF1 | 10522 | 8 | 38 | 50 |
Total number of gains: 27; Total number of losses: 120; Total Number of normals: 341.
Somatic mutations of DEAF1:
Generating mutation plots.
Highly correlated genes for DEAF1:
Showing top 20/1158 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DEAF1 | ZNF823 | 0.738661 | 4 | 0 | 4 |
| DEAF1 | CRIP1 | 0.711168 | 3 | 0 | 3 |
| DEAF1 | RNF214 | 0.705889 | 3 | 0 | 3 |
| DEAF1 | CRYL1 | 0.696163 | 10 | 0 | 10 |
| DEAF1 | GTPBP2 | 0.693652 | 6 | 0 | 6 |
| DEAF1 | ZNF554 | 0.693273 | 3 | 0 | 3 |
| DEAF1 | EXOC4 | 0.690585 | 3 | 0 | 3 |
| DEAF1 | KCTD21 | 0.686386 | 4 | 0 | 3 |
| DEAF1 | PPFIBP2 | 0.682522 | 11 | 0 | 10 |
| DEAF1 | NFIX | 0.680709 | 4 | 0 | 4 |
| DEAF1 | KIF21A | 0.680246 | 3 | 0 | 3 |
| DEAF1 | NDUFB3 | 0.680131 | 3 | 0 | 3 |
| DEAF1 | CUL3 | 0.678911 | 10 | 0 | 9 |
| DEAF1 | YPEL3 | 0.67878 | 7 | 0 | 6 |
| DEAF1 | RNH1 | 0.677321 | 11 | 0 | 11 |
| DEAF1 | ZNF506 | 0.677074 | 3 | 0 | 3 |
| DEAF1 | TSG101 | 0.676854 | 12 | 0 | 11 |
| DEAF1 | GALNT12 | 0.67652 | 11 | 0 | 10 |
| DEAF1 | GPR108 | 0.675437 | 3 | 0 | 3 |
| DEAF1 | ACER3 | 0.675018 | 3 | 0 | 3 |
For details and further investigation, click here