| Full name: cysteine rich protein 1 | Alias Symbol: CRIP | ||
| Type: protein-coding gene | Cytoband: 14q32.33 | ||
| Entrez ID: 1396 | HGNC ID: HGNC:2360 | Ensembl Gene: ENSG00000213145 | OMIM ID: 123875 |
| Drug and gene relationship at DGIdb | |||
Expression of CRIP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CRIP1 | 1396 | 205081_at | -0.0502 | 0.9516 | |
| GSE20347 | CRIP1 | 1396 | 205081_at | -1.3648 | 0.0003 | |
| GSE23400 | CRIP1 | 1396 | 205081_at | -0.5285 | 0.0108 | |
| GSE26886 | CRIP1 | 1396 | 205081_at | 0.9397 | 0.0878 | |
| GSE29001 | CRIP1 | 1396 | 205081_at | -0.4340 | 0.5397 | |
| GSE38129 | CRIP1 | 1396 | 205081_at | -1.3538 | 0.0001 | |
| GSE45670 | CRIP1 | 1396 | 205081_at | -0.7410 | 0.0478 | |
| GSE53622 | CRIP1 | 1396 | 45866 | 0.4431 | 0.0070 | |
| GSE53624 | CRIP1 | 1396 | 45866 | -0.2644 | 0.0125 | |
| GSE63941 | CRIP1 | 1396 | 205081_at | 1.9642 | 0.3532 | |
| GSE77861 | CRIP1 | 1396 | 205081_at | -0.3786 | 0.6337 | |
| GSE97050 | CRIP1 | 1396 | A_33_P3251703 | 0.3026 | 0.5234 | |
| SRP007169 | CRIP1 | 1396 | RNAseq | -0.8496 | 0.1395 | |
| SRP008496 | CRIP1 | 1396 | RNAseq | -0.9246 | 0.0175 | |
| SRP064894 | CRIP1 | 1396 | RNAseq | 0.5092 | 0.3372 | |
| SRP133303 | CRIP1 | 1396 | RNAseq | -0.9480 | 0.0116 | |
| SRP159526 | CRIP1 | 1396 | RNAseq | -1.2851 | 0.0001 | |
| SRP193095 | CRIP1 | 1396 | RNAseq | -0.5930 | 0.0639 | |
| SRP219564 | CRIP1 | 1396 | RNAseq | 0.2905 | 0.6595 | |
| TCGA | CRIP1 | 1396 | RNAseq | -0.1880 | 0.1880 |
Upregulated datasets: 0; Downregulated datasets: 3.
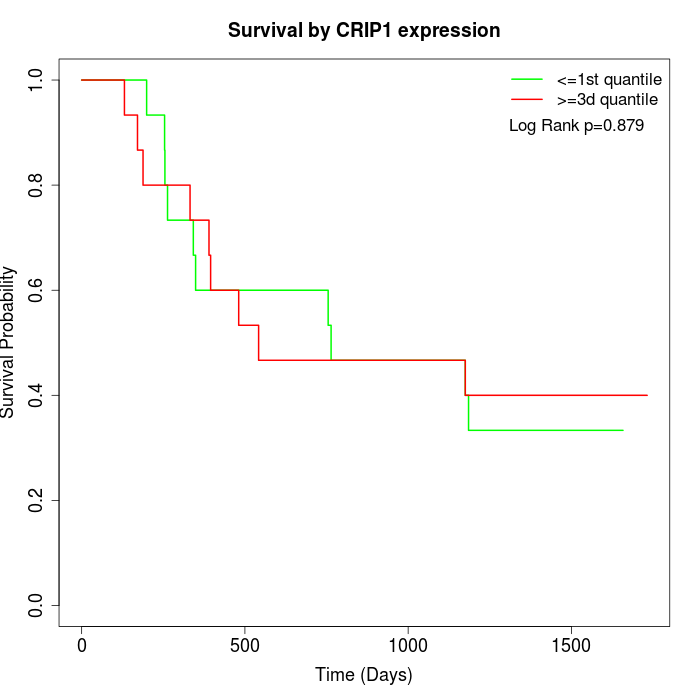
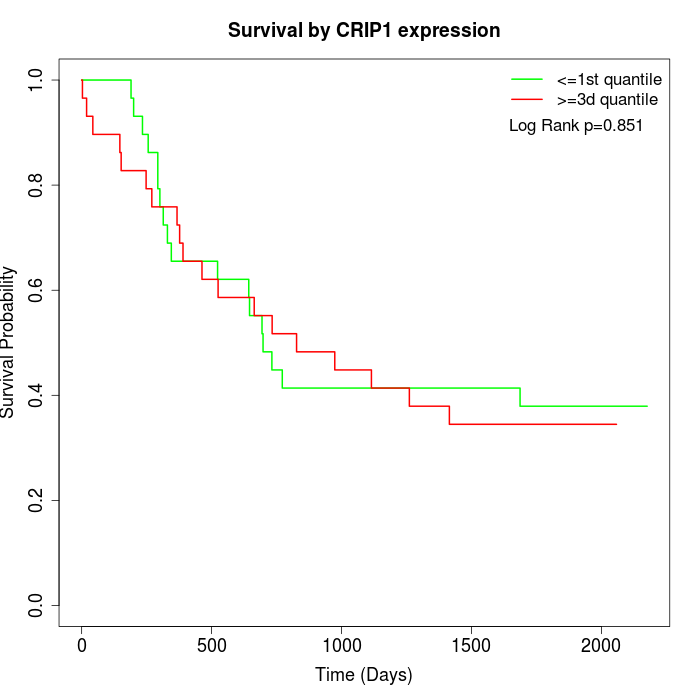
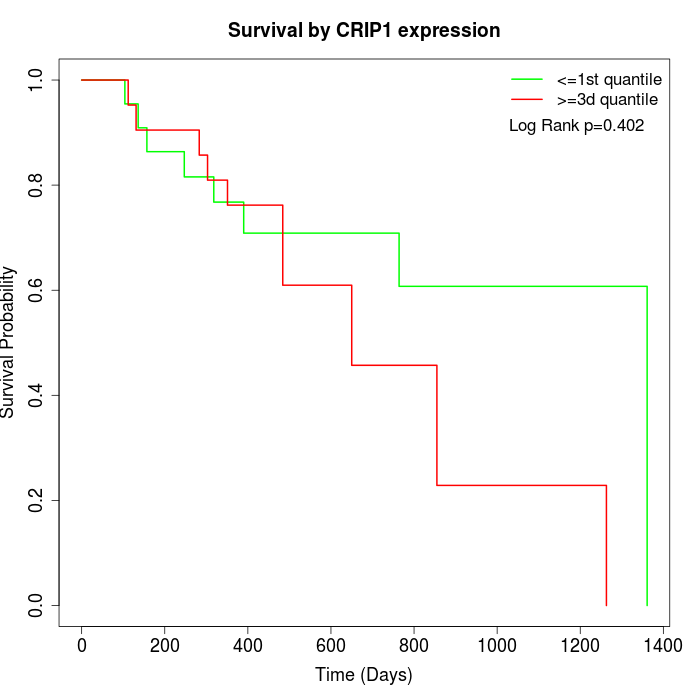
Survival by CRIP1 expression:
Note: Click image to view full size file.
Copy number change of CRIP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CRIP1 | 1396 | 10 | 3 | 17 | |
| GSE20123 | CRIP1 | 1396 | 10 | 2 | 18 | |
| GSE43470 | CRIP1 | 1396 | 5 | 3 | 35 | |
| GSE46452 | CRIP1 | 1396 | 18 | 4 | 37 | |
| GSE47630 | CRIP1 | 1396 | 10 | 10 | 20 | |
| GSE54993 | CRIP1 | 1396 | 4 | 11 | 55 | |
| GSE54994 | CRIP1 | 1396 | 21 | 4 | 28 | |
| GSE60625 | CRIP1 | 1396 | 0 | 2 | 9 | |
| GSE74703 | CRIP1 | 1396 | 4 | 3 | 29 | |
| GSE74704 | CRIP1 | 1396 | 5 | 2 | 13 | |
| TCGA | CRIP1 | 1396 | 31 | 18 | 47 |
Total number of gains: 118; Total number of losses: 62; Total Number of normals: 308.
Somatic mutations of CRIP1:
Generating mutation plots.
Highly correlated genes for CRIP1:
Showing top 20/129 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CRIP1 | PPP2R5C | 0.78128 | 4 | 0 | 4 |
| CRIP1 | ERC1 | 0.720467 | 3 | 0 | 3 |
| CRIP1 | LIMCH1 | 0.715485 | 3 | 0 | 3 |
| CRIP1 | COL14A1 | 0.712145 | 3 | 0 | 3 |
| CRIP1 | AP4M1 | 0.711509 | 3 | 0 | 3 |
| CRIP1 | DEAF1 | 0.711168 | 3 | 0 | 3 |
| CRIP1 | OXTR | 0.695251 | 3 | 0 | 3 |
| CRIP1 | HLCS | 0.687551 | 4 | 0 | 3 |
| CRIP1 | SLC12A4 | 0.684456 | 3 | 0 | 3 |
| CRIP1 | PILRA | 0.683994 | 3 | 0 | 3 |
| CRIP1 | DUSP22 | 0.681658 | 3 | 0 | 3 |
| CRIP1 | TSC22D3 | 0.67936 | 3 | 0 | 3 |
| CRIP1 | TMEM25 | 0.678068 | 3 | 0 | 3 |
| CRIP1 | AFF1 | 0.664783 | 4 | 0 | 4 |
| CRIP1 | KLHL2 | 0.664619 | 3 | 0 | 3 |
| CRIP1 | PITPNM2 | 0.657681 | 3 | 0 | 3 |
| CRIP1 | GIMAP6 | 0.655485 | 4 | 0 | 4 |
| CRIP1 | FAM114A2 | 0.646982 | 3 | 0 | 3 |
| CRIP1 | GNE | 0.643253 | 3 | 0 | 3 |
| CRIP1 | METTL7A | 0.642896 | 3 | 0 | 3 |
For details and further investigation, click here