| Full name: dystonin | Alias Symbol: BP240|KIAA0728|FLJ21489|FLJ13425|FLJ32235|FLJ30627|CATX-15|BPA|MACF2 | ||
| Type: protein-coding gene | Cytoband: 6p12.1 | ||
| Entrez ID: 667 | HGNC ID: HGNC:1090 | Ensembl Gene: ENSG00000151914 | OMIM ID: 113810 |
| Drug and gene relationship at DGIdb | |||
Expression of DST:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DST | 667 | 204455_at | 1.0281 | 0.1406 | |
| GSE20347 | DST | 667 | 204455_at | 0.3829 | 0.3766 | |
| GSE23400 | DST | 667 | 216918_s_at | 1.1175 | 0.0000 | |
| GSE26886 | DST | 667 | 204455_at | 0.6551 | 0.0284 | |
| GSE29001 | DST | 667 | 204455_at | 0.1119 | 0.8511 | |
| GSE38129 | DST | 667 | 204455_at | 1.2256 | 0.0798 | |
| GSE45670 | DST | 667 | 204455_at | 0.6710 | 0.0689 | |
| GSE53622 | DST | 667 | 51313 | -0.0942 | 0.4575 | |
| GSE53624 | DST | 667 | 51313 | 0.1518 | 0.1028 | |
| GSE63941 | DST | 667 | 215016_x_at | -1.8530 | 0.0045 | |
| GSE77861 | DST | 667 | 204455_at | 0.8525 | 0.0884 | |
| GSE97050 | DST | 667 | A_23_P303718 | -0.6704 | 0.1754 | |
| SRP007169 | DST | 667 | RNAseq | 1.9408 | 0.0000 | |
| SRP008496 | DST | 667 | RNAseq | 2.0630 | 0.0000 | |
| SRP064894 | DST | 667 | RNAseq | 0.3009 | 0.3974 | |
| SRP133303 | DST | 667 | RNAseq | 0.7712 | 0.0004 | |
| SRP159526 | DST | 667 | RNAseq | -0.3835 | 0.4689 | |
| SRP193095 | DST | 667 | RNAseq | 0.6596 | 0.0088 | |
| SRP219564 | DST | 667 | RNAseq | -0.0948 | 0.8577 | |
| TCGA | DST | 667 | RNAseq | 0.0711 | 0.3630 |
Upregulated datasets: 3; Downregulated datasets: 1.
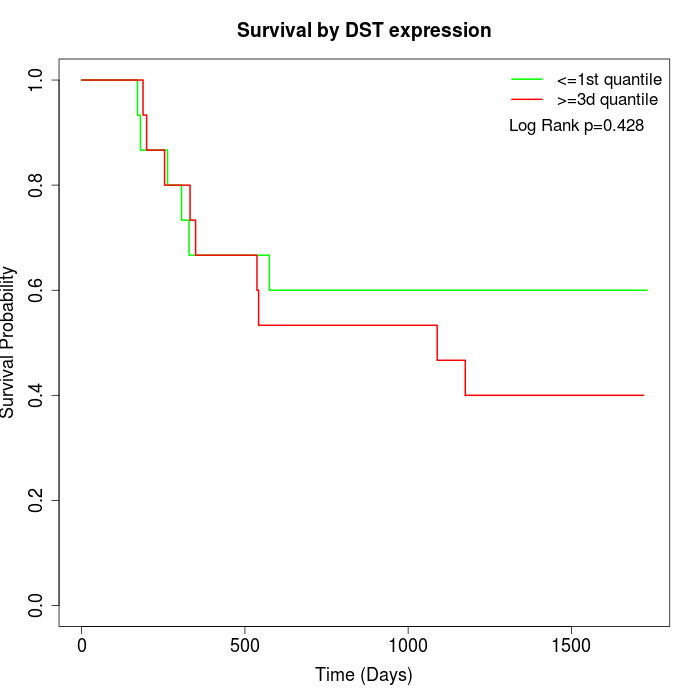
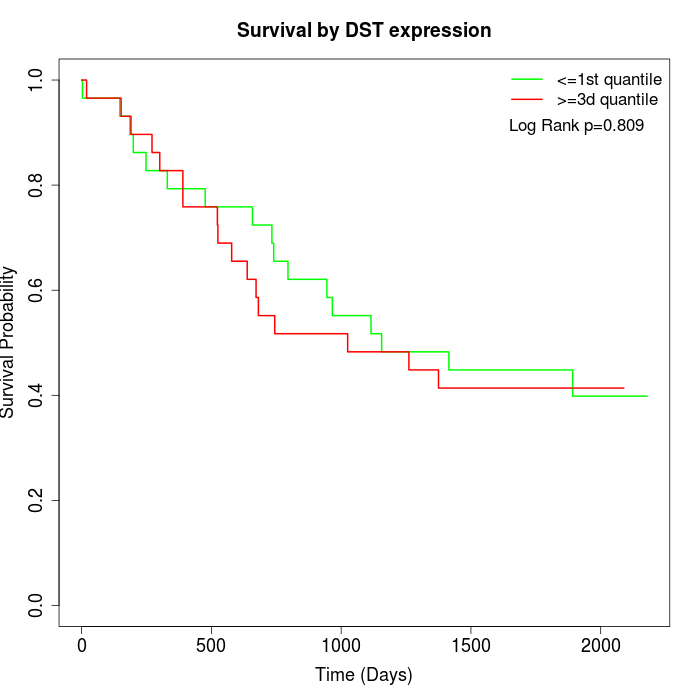
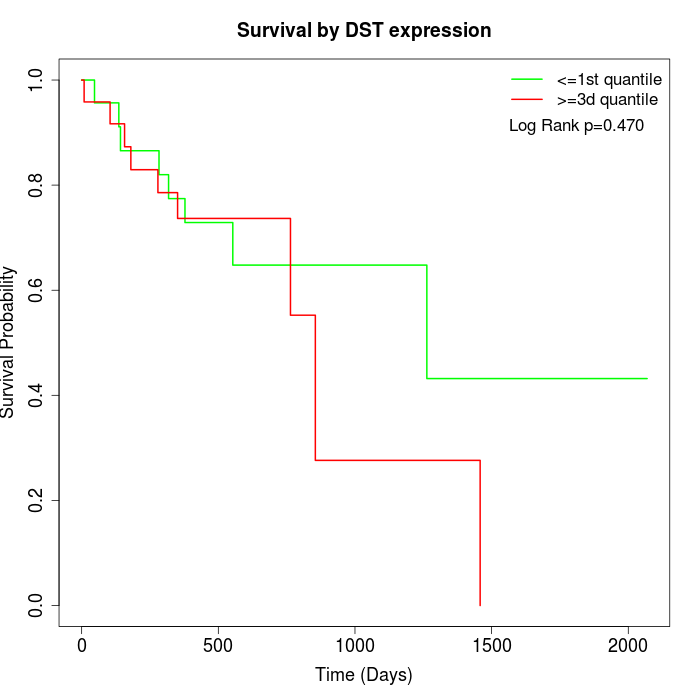
Survival by DST expression:
Note: Click image to view full size file.
Copy number change of DST:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DST | 667 | 9 | 1 | 20 | |
| GSE20123 | DST | 667 | 9 | 1 | 20 | |
| GSE43470 | DST | 667 | 5 | 0 | 38 | |
| GSE46452 | DST | 667 | 2 | 10 | 47 | |
| GSE47630 | DST | 667 | 7 | 5 | 28 | |
| GSE54993 | DST | 667 | 3 | 2 | 65 | |
| GSE54994 | DST | 667 | 9 | 4 | 40 | |
| GSE60625 | DST | 667 | 0 | 1 | 10 | |
| GSE74703 | DST | 667 | 5 | 0 | 31 | |
| GSE74704 | DST | 667 | 5 | 0 | 15 | |
| TCGA | DST | 667 | 23 | 11 | 62 |
Total number of gains: 77; Total number of losses: 35; Total Number of normals: 376.
Somatic mutations of DST:
Generating mutation plots.
Highly correlated genes for DST:
Showing top 20/241 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DST | ZEB1 | 0.702102 | 3 | 0 | 3 |
| DST | DPH7 | 0.678076 | 3 | 0 | 3 |
| DST | ADAMTS12 | 0.675547 | 3 | 0 | 3 |
| DST | EBLN2 | 0.670965 | 3 | 0 | 3 |
| DST | GTPBP6 | 0.663322 | 3 | 0 | 3 |
| DST | SYT14 | 0.659014 | 3 | 0 | 3 |
| DST | C4orf48 | 0.649304 | 3 | 0 | 3 |
| DST | LOXL2 | 0.647508 | 3 | 0 | 3 |
| DST | PHF20L1 | 0.640257 | 5 | 0 | 3 |
| DST | FJX1 | 0.637863 | 4 | 0 | 4 |
| DST | NIPBL | 0.635638 | 3 | 0 | 3 |
| DST | SFXN5 | 0.634491 | 3 | 0 | 3 |
| DST | TATDN1 | 0.630579 | 4 | 0 | 4 |
| DST | METTL8 | 0.62831 | 5 | 0 | 4 |
| DST | DDX31 | 0.626816 | 5 | 0 | 4 |
| DST | MFAP2 | 0.625604 | 4 | 0 | 3 |
| DST | KPNA1 | 0.616106 | 5 | 0 | 4 |
| DST | HNRNPA1 | 0.614094 | 3 | 0 | 3 |
| DST | LZTR1 | 0.613681 | 3 | 0 | 3 |
| DST | CIR1 | 0.612515 | 3 | 0 | 3 |
For details and further investigation, click here