| Full name: dual specificity phosphatase 5 | Alias Symbol: HVH3 | ||
| Type: protein-coding gene | Cytoband: 10q25.2 | ||
| Entrez ID: 1847 | HGNC ID: HGNC:3071 | Ensembl Gene: ENSG00000138166 | OMIM ID: 603069 |
| Drug and gene relationship at DGIdb | |||
DUSP5 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway |
Expression of DUSP5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DUSP5 | 1847 | 209457_at | -1.4895 | 0.1752 | |
| GSE20347 | DUSP5 | 1847 | 209457_at | -2.6713 | 0.0000 | |
| GSE23400 | DUSP5 | 1847 | 209457_at | -2.3531 | 0.0000 | |
| GSE26886 | DUSP5 | 1847 | 209457_at | -4.3295 | 0.0000 | |
| GSE29001 | DUSP5 | 1847 | 209457_at | -2.7422 | 0.0000 | |
| GSE38129 | DUSP5 | 1847 | 209457_at | -2.2340 | 0.0000 | |
| GSE45670 | DUSP5 | 1847 | 209457_at | -1.1803 | 0.0177 | |
| GSE53622 | DUSP5 | 1847 | 65521 | -2.5300 | 0.0000 | |
| GSE53624 | DUSP5 | 1847 | 65521 | -2.8181 | 0.0000 | |
| GSE63941 | DUSP5 | 1847 | 209457_at | 0.0388 | 0.9767 | |
| GSE77861 | DUSP5 | 1847 | 209457_at | -1.6137 | 0.0155 | |
| GSE97050 | DUSP5 | 1847 | A_23_P150018 | -0.6794 | 0.4183 | |
| SRP007169 | DUSP5 | 1847 | RNAseq | -5.2999 | 0.0000 | |
| SRP008496 | DUSP5 | 1847 | RNAseq | -5.1573 | 0.0000 | |
| SRP064894 | DUSP5 | 1847 | RNAseq | -3.6277 | 0.0000 | |
| SRP133303 | DUSP5 | 1847 | RNAseq | -2.2120 | 0.0000 | |
| SRP159526 | DUSP5 | 1847 | RNAseq | -3.0104 | 0.0000 | |
| SRP193095 | DUSP5 | 1847 | RNAseq | -3.0739 | 0.0000 | |
| SRP219564 | DUSP5 | 1847 | RNAseq | -1.9696 | 0.0058 | |
| TCGA | DUSP5 | 1847 | RNAseq | -0.0006 | 0.9964 |
Upregulated datasets: 0; Downregulated datasets: 16.
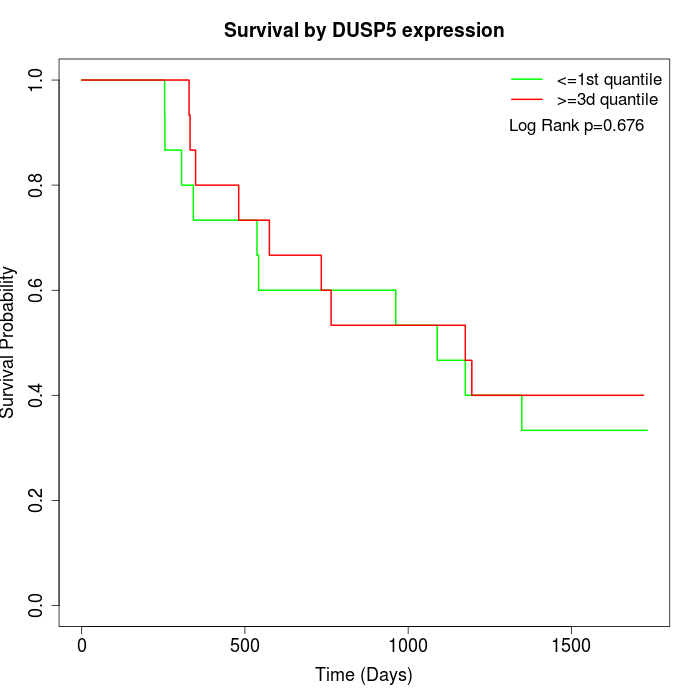
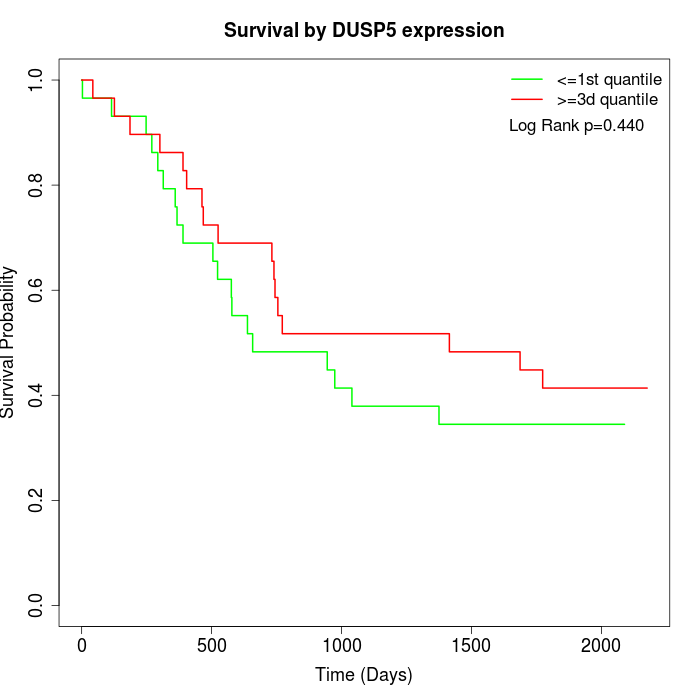
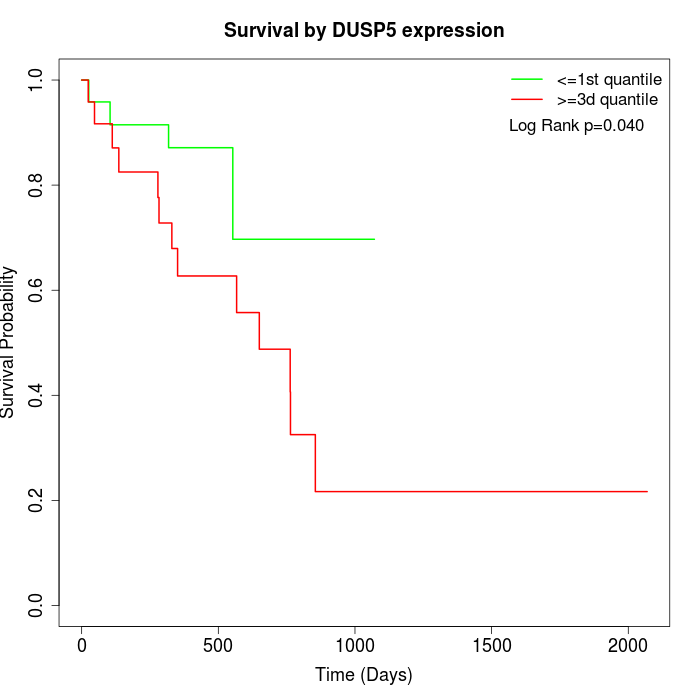
Survival by DUSP5 expression:
Note: Click image to view full size file.
Copy number change of DUSP5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DUSP5 | 1847 | 1 | 10 | 19 | |
| GSE20123 | DUSP5 | 1847 | 1 | 9 | 20 | |
| GSE43470 | DUSP5 | 1847 | 0 | 7 | 36 | |
| GSE46452 | DUSP5 | 1847 | 0 | 11 | 48 | |
| GSE47630 | DUSP5 | 1847 | 2 | 14 | 24 | |
| GSE54993 | DUSP5 | 1847 | 8 | 0 | 62 | |
| GSE54994 | DUSP5 | 1847 | 1 | 10 | 42 | |
| GSE60625 | DUSP5 | 1847 | 0 | 0 | 11 | |
| GSE74703 | DUSP5 | 1847 | 0 | 5 | 31 | |
| GSE74704 | DUSP5 | 1847 | 1 | 4 | 15 | |
| TCGA | DUSP5 | 1847 | 4 | 28 | 64 |
Total number of gains: 18; Total number of losses: 98; Total Number of normals: 372.
Somatic mutations of DUSP5:
Generating mutation plots.
Highly correlated genes for DUSP5:
Showing top 20/1602 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DUSP5 | TRIP10 | 0.856922 | 11 | 0 | 11 |
| DUSP5 | C18orf25 | 0.854223 | 11 | 0 | 11 |
| DUSP5 | RANBP9 | 0.846503 | 11 | 0 | 11 |
| DUSP5 | RAET1E | 0.844522 | 6 | 0 | 6 |
| DUSP5 | ENDOU | 0.836339 | 11 | 0 | 11 |
| DUSP5 | SCNN1B | 0.835311 | 10 | 0 | 10 |
| DUSP5 | SPINK7 | 0.834941 | 6 | 0 | 6 |
| DUSP5 | IL36A | 0.834248 | 10 | 0 | 10 |
| DUSP5 | EHD3 | 0.833399 | 11 | 0 | 11 |
| DUSP5 | YOD1 | 0.832751 | 11 | 0 | 11 |
| DUSP5 | SCEL | 0.831735 | 10 | 0 | 10 |
| DUSP5 | PMM1 | 0.827111 | 10 | 0 | 10 |
| DUSP5 | SASH1 | 0.825921 | 11 | 0 | 11 |
| DUSP5 | VASN | 0.825901 | 7 | 0 | 7 |
| DUSP5 | PIM1 | 0.825042 | 11 | 0 | 11 |
| DUSP5 | PADI1 | 0.824138 | 8 | 0 | 8 |
| DUSP5 | TTC9 | 0.823792 | 11 | 0 | 11 |
| DUSP5 | CDKN2AIP | 0.823358 | 11 | 0 | 11 |
| DUSP5 | SLURP1 | 0.822792 | 10 | 0 | 10 |
| DUSP5 | MPZL3 | 0.821638 | 7 | 0 | 7 |
For details and further investigation, click here