| Full name: dual specificity phosphatase 1 | Alias Symbol: HVH1|CL100|MKP-1 | ||
| Type: protein-coding gene | Cytoband: 5q35.1 | ||
| Entrez ID: 1843 | HGNC ID: HGNC:3064 | Ensembl Gene: ENSG00000120129 | OMIM ID: 600714 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
DUSP1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04726 | Serotonergic synapse |
Expression of DUSP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DUSP1 | 1843 | 201041_s_at | -0.6398 | 0.3802 | |
| GSE20347 | DUSP1 | 1843 | 201041_s_at | -1.5477 | 0.0000 | |
| GSE23400 | DUSP1 | 1843 | 201041_s_at | -1.0220 | 0.0000 | |
| GSE26886 | DUSP1 | 1843 | 201041_s_at | -0.0153 | 0.9682 | |
| GSE29001 | DUSP1 | 1843 | 201041_s_at | -1.9437 | 0.0000 | |
| GSE38129 | DUSP1 | 1843 | 201041_s_at | -1.7600 | 0.0000 | |
| GSE45670 | DUSP1 | 1843 | 201041_s_at | -1.3640 | 0.0005 | |
| GSE53622 | DUSP1 | 1843 | 82420 | -1.6541 | 0.0000 | |
| GSE53624 | DUSP1 | 1843 | 82420 | -1.8160 | 0.0000 | |
| GSE63941 | DUSP1 | 1843 | 201041_s_at | -1.9763 | 0.0679 | |
| GSE77861 | DUSP1 | 1843 | 201041_s_at | -0.5110 | 0.4590 | |
| GSE97050 | DUSP1 | 1843 | A_23_P110712 | -0.5802 | 0.2840 | |
| SRP007169 | DUSP1 | 1843 | RNAseq | -3.8418 | 0.0000 | |
| SRP008496 | DUSP1 | 1843 | RNAseq | -3.3898 | 0.0000 | |
| SRP064894 | DUSP1 | 1843 | RNAseq | -1.3879 | 0.0000 | |
| SRP133303 | DUSP1 | 1843 | RNAseq | -0.7984 | 0.0367 | |
| SRP159526 | DUSP1 | 1843 | RNAseq | -0.8119 | 0.1961 | |
| SRP193095 | DUSP1 | 1843 | RNAseq | -0.1932 | 0.6070 | |
| SRP219564 | DUSP1 | 1843 | RNAseq | 0.2282 | 0.5389 | |
| TCGA | DUSP1 | 1843 | RNAseq | -0.2110 | 0.0268 |
Upregulated datasets: 0; Downregulated datasets: 10.
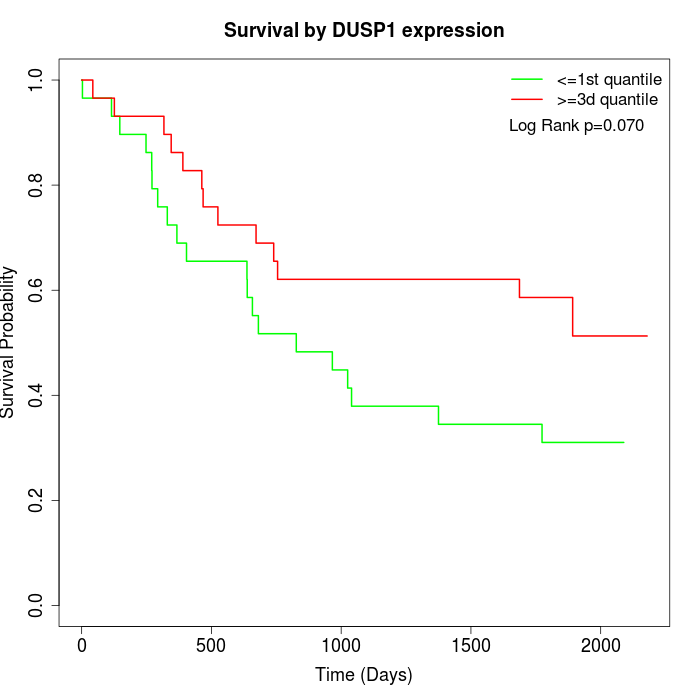
Survival by DUSP1 expression:
Note: Click image to view full size file.
Copy number change of DUSP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DUSP1 | 1843 | 2 | 13 | 15 | |
| GSE20123 | DUSP1 | 1843 | 2 | 13 | 15 | |
| GSE43470 | DUSP1 | 1843 | 1 | 10 | 32 | |
| GSE46452 | DUSP1 | 1843 | 0 | 27 | 32 | |
| GSE47630 | DUSP1 | 1843 | 0 | 20 | 20 | |
| GSE54993 | DUSP1 | 1843 | 9 | 2 | 59 | |
| GSE54994 | DUSP1 | 1843 | 2 | 15 | 36 | |
| GSE60625 | DUSP1 | 1843 | 1 | 0 | 10 | |
| GSE74703 | DUSP1 | 1843 | 1 | 7 | 28 | |
| GSE74704 | DUSP1 | 1843 | 1 | 6 | 13 | |
| TCGA | DUSP1 | 1843 | 8 | 34 | 54 |
Total number of gains: 27; Total number of losses: 147; Total Number of normals: 314.
Somatic mutations of DUSP1:
Generating mutation plots.
Highly correlated genes for DUSP1:
Showing top 20/671 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DUSP1 | DUSP2 | 0.813199 | 4 | 0 | 4 |
| DUSP1 | FOS | 0.805154 | 12 | 0 | 12 |
| DUSP1 | ZFP36 | 0.762575 | 11 | 0 | 10 |
| DUSP1 | PRDM8 | 0.743718 | 3 | 0 | 3 |
| DUSP1 | JUNB | 0.731374 | 11 | 0 | 11 |
| DUSP1 | CSRNP1 | 0.71783 | 7 | 0 | 7 |
| DUSP1 | ZNF460 | 0.702224 | 3 | 0 | 3 |
| DUSP1 | CYP21A2 | 0.691567 | 3 | 0 | 3 |
| DUSP1 | KLF6 | 0.688833 | 12 | 0 | 10 |
| DUSP1 | TOB1 | 0.67975 | 11 | 0 | 10 |
| DUSP1 | SPHKAP | 0.678956 | 4 | 0 | 4 |
| DUSP1 | RASSF3 | 0.677441 | 4 | 0 | 3 |
| DUSP1 | EGR1 | 0.675648 | 12 | 0 | 10 |
| DUSP1 | SORBS2 | 0.674343 | 8 | 0 | 8 |
| DUSP1 | BHLHE40 | 0.673206 | 10 | 0 | 9 |
| DUSP1 | FOSB | 0.670796 | 13 | 0 | 12 |
| DUSP1 | KLF4 | 0.670255 | 10 | 0 | 9 |
| DUSP1 | PLAC9 | 0.669032 | 4 | 0 | 4 |
| DUSP1 | MIDN | 0.667609 | 6 | 0 | 5 |
| DUSP1 | KCNB1 | 0.66754 | 3 | 0 | 3 |
For details and further investigation, click here