| Full name: euchromatic histone lysine methyltransferase 1 | Alias Symbol: Eu-HMTase1|FLJ12879|KIAA1876|bA188C12.1|KMT1D|FLJ40292 | ||
| Type: protein-coding gene | Cytoband: 9q34.3 | ||
| Entrez ID: 79813 | HGNC ID: HGNC:24650 | Ensembl Gene: ENSG00000181090 | OMIM ID: 607001 |
| Drug and gene relationship at DGIdb | |||
EHMT1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04211 | Longevity regulating pathway - mammal |
Expression of EHMT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EHMT1 | 79813 | 225461_at | 0.0494 | 0.9417 | |
| GSE20347 | EHMT1 | 79813 | 219339_s_at | -0.0791 | 0.3965 | |
| GSE23400 | EHMT1 | 79813 | 219339_s_at | -0.0526 | 0.1707 | |
| GSE26886 | EHMT1 | 79813 | 225461_at | 0.5427 | 0.0303 | |
| GSE29001 | EHMT1 | 79813 | 219339_s_at | 0.0184 | 0.9218 | |
| GSE38129 | EHMT1 | 79813 | 219339_s_at | -0.0709 | 0.4065 | |
| GSE45670 | EHMT1 | 79813 | 225461_at | 0.3107 | 0.0312 | |
| GSE53622 | EHMT1 | 79813 | 10399 | 0.2628 | 0.0005 | |
| GSE53624 | EHMT1 | 79813 | 10399 | 0.2274 | 0.0004 | |
| GSE63941 | EHMT1 | 79813 | 225461_at | -0.0223 | 0.9624 | |
| GSE77861 | EHMT1 | 79813 | 225461_at | 0.4882 | 0.0357 | |
| GSE97050 | EHMT1 | 79813 | A_33_P3286387 | 0.2355 | 0.2819 | |
| SRP007169 | EHMT1 | 79813 | RNAseq | 0.7584 | 0.0225 | |
| SRP008496 | EHMT1 | 79813 | RNAseq | 0.6176 | 0.0053 | |
| SRP064894 | EHMT1 | 79813 | RNAseq | 0.0881 | 0.5532 | |
| SRP133303 | EHMT1 | 79813 | RNAseq | 0.1248 | 0.3104 | |
| SRP159526 | EHMT1 | 79813 | RNAseq | 0.3086 | 0.1065 | |
| SRP193095 | EHMT1 | 79813 | RNAseq | 0.3309 | 0.0144 | |
| SRP219564 | EHMT1 | 79813 | RNAseq | -0.0841 | 0.7026 | |
| TCGA | EHMT1 | 79813 | RNAseq | 0.0181 | 0.6901 |
Upregulated datasets: 0; Downregulated datasets: 0.
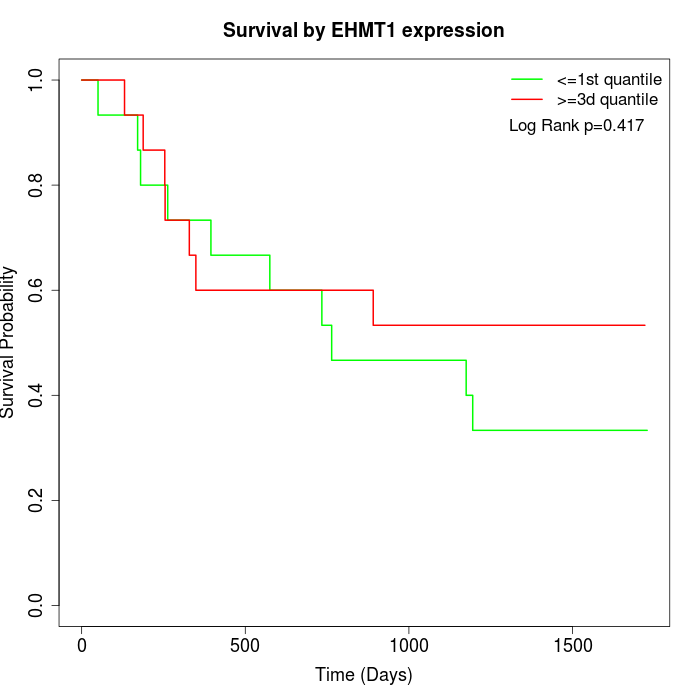
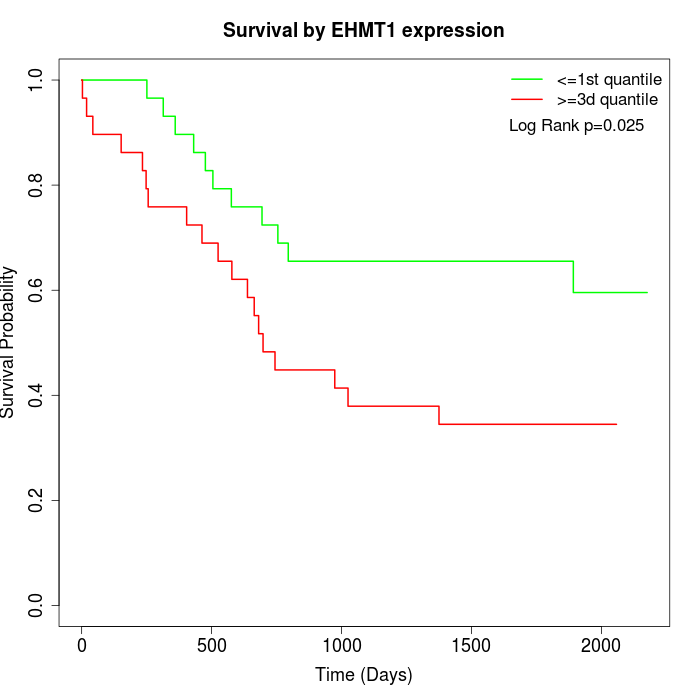
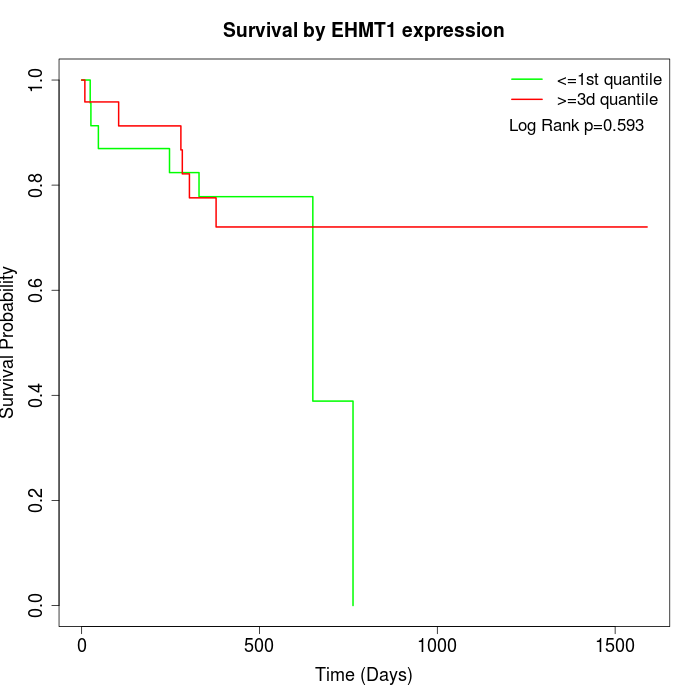
Survival by EHMT1 expression:
Note: Click image to view full size file.
Copy number change of EHMT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EHMT1 | 79813 | 6 | 7 | 17 | |
| GSE20123 | EHMT1 | 79813 | 6 | 7 | 17 | |
| GSE43470 | EHMT1 | 79813 | 3 | 7 | 33 | |
| GSE46452 | EHMT1 | 79813 | 6 | 13 | 40 | |
| GSE47630 | EHMT1 | 79813 | 6 | 15 | 19 | |
| GSE54993 | EHMT1 | 79813 | 3 | 3 | 64 | |
| GSE54994 | EHMT1 | 79813 | 19 | 3 | 31 | |
| GSE60625 | EHMT1 | 79813 | 0 | 0 | 11 | |
| GSE74703 | EHMT1 | 79813 | 3 | 5 | 28 | |
| GSE74704 | EHMT1 | 79813 | 3 | 5 | 12 | |
| TCGA | EHMT1 | 79813 | 28 | 24 | 44 |
Total number of gains: 83; Total number of losses: 89; Total Number of normals: 316.
Somatic mutations of EHMT1:
Generating mutation plots.
Highly correlated genes for EHMT1:
Showing top 20/96 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EHMT1 | CYP2S1 | 0.676588 | 3 | 0 | 3 |
| EHMT1 | MS4A1 | 0.66544 | 3 | 0 | 3 |
| EHMT1 | ATP1B3 | 0.637294 | 3 | 0 | 3 |
| EHMT1 | NELFB | 0.635357 | 7 | 0 | 6 |
| EHMT1 | TNS4 | 0.633546 | 3 | 0 | 3 |
| EHMT1 | SCYL1 | 0.632657 | 3 | 0 | 3 |
| EHMT1 | HOXB7 | 0.630322 | 3 | 0 | 3 |
| EHMT1 | MAFK | 0.629785 | 3 | 0 | 3 |
| EHMT1 | SUGP2 | 0.619735 | 3 | 0 | 3 |
| EHMT1 | SPINT2 | 0.61936 | 3 | 0 | 3 |
| EHMT1 | DCAF7 | 0.615297 | 3 | 0 | 3 |
| EHMT1 | B4GALT7 | 0.610827 | 4 | 0 | 3 |
| EHMT1 | SPPL2B | 0.606508 | 5 | 0 | 4 |
| EHMT1 | CBFA2T2 | 0.605486 | 3 | 0 | 3 |
| EHMT1 | ZNF574 | 0.598508 | 4 | 0 | 4 |
| EHMT1 | SHKBP1 | 0.598297 | 5 | 0 | 3 |
| EHMT1 | TMEM203 | 0.597579 | 8 | 0 | 6 |
| EHMT1 | SULT1A2 | 0.597522 | 3 | 0 | 3 |
| EHMT1 | NOL8 | 0.596618 | 6 | 0 | 5 |
| EHMT1 | ULK1 | 0.585857 | 4 | 0 | 3 |
For details and further investigation, click here