| Full name: ceramide synthase 5 | Alias Symbol: Trh4|MGC45411|FLJ25304 | ||
| Type: protein-coding gene | Cytoband: 12q13.12 | ||
| Entrez ID: 91012 | HGNC ID: HGNC:23749 | Ensembl Gene: ENSG00000139624 | OMIM ID: 615335 |
| Drug and gene relationship at DGIdb | |||
Expression of CERS5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CERS5 | 91012 | 224951_at | 0.3413 | 0.1427 | |
| GSE26886 | CERS5 | 91012 | 224951_at | 0.2700 | 0.1793 | |
| GSE45670 | CERS5 | 91012 | 224951_at | 0.0617 | 0.5908 | |
| GSE53622 | CERS5 | 91012 | 34678 | 0.3395 | 0.0000 | |
| GSE53624 | CERS5 | 91012 | 34678 | 0.6104 | 0.0000 | |
| GSE63941 | CERS5 | 91012 | 224951_at | -0.2414 | 0.3731 | |
| GSE77861 | CERS5 | 91012 | 224951_at | 0.4724 | 0.0041 | |
| GSE97050 | CERS5 | 91012 | A_33_P3285038 | 0.1597 | 0.5313 | |
| SRP007169 | CERS5 | 91012 | RNAseq | 0.3990 | 0.3033 | |
| SRP008496 | CERS5 | 91012 | RNAseq | 0.7063 | 0.0032 | |
| SRP064894 | CERS5 | 91012 | RNAseq | 0.6278 | 0.0003 | |
| SRP133303 | CERS5 | 91012 | RNAseq | 0.3128 | 0.0001 | |
| SRP159526 | CERS5 | 91012 | RNAseq | 0.5519 | 0.1011 | |
| SRP193095 | CERS5 | 91012 | RNAseq | 0.4248 | 0.0000 | |
| SRP219564 | CERS5 | 91012 | RNAseq | 0.1765 | 0.4823 |
Upregulated datasets: 0; Downregulated datasets: 0.
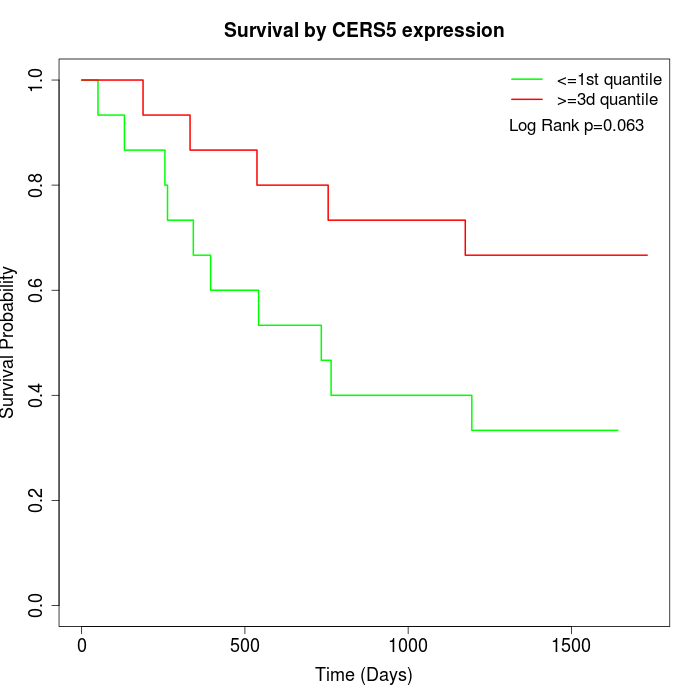
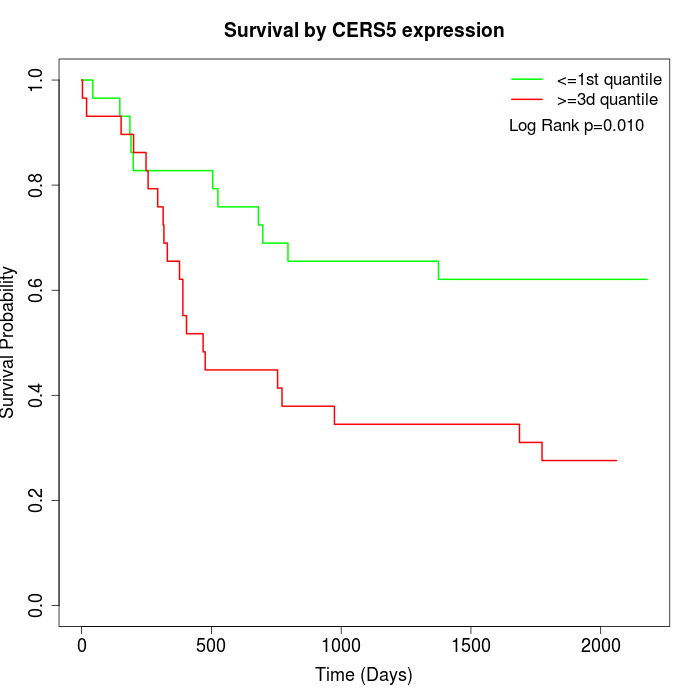
Survival by CERS5 expression:
Note: Click image to view full size file.
Copy number change of CERS5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CERS5 | 91012 | 8 | 1 | 21 | |
| GSE20123 | CERS5 | 91012 | 8 | 1 | 21 | |
| GSE43470 | CERS5 | 91012 | 3 | 0 | 40 | |
| GSE46452 | CERS5 | 91012 | 8 | 1 | 50 | |
| GSE47630 | CERS5 | 91012 | 11 | 2 | 27 | |
| GSE54993 | CERS5 | 91012 | 0 | 5 | 65 | |
| GSE54994 | CERS5 | 91012 | 4 | 1 | 48 | |
| GSE60625 | CERS5 | 91012 | 0 | 0 | 11 | |
| GSE74703 | CERS5 | 91012 | 3 | 0 | 33 | |
| GSE74704 | CERS5 | 91012 | 5 | 1 | 14 | |
| TCGA | CERS5 | 91012 | 15 | 11 | 70 |
Total number of gains: 65; Total number of losses: 23; Total Number of normals: 400.
Somatic mutations of CERS5:
Generating mutation plots.
Highly correlated genes for CERS5:
Showing top 20/1068 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CERS5 | CDC42EP4 | 0.832278 | 3 | 0 | 3 |
| CERS5 | GGNBP2 | 0.786075 | 3 | 0 | 3 |
| CERS5 | KLF11 | 0.766258 | 3 | 0 | 3 |
| CERS5 | SDHAF2 | 0.754326 | 3 | 0 | 3 |
| CERS5 | FAM89B | 0.752046 | 3 | 0 | 3 |
| CERS5 | BCL7A | 0.747728 | 5 | 0 | 5 |
| CERS5 | UROS | 0.746591 | 3 | 0 | 3 |
| CERS5 | ELAVL1 | 0.741755 | 3 | 0 | 3 |
| CERS5 | POMZP3 | 0.741465 | 3 | 0 | 3 |
| CERS5 | CCDC127 | 0.727961 | 3 | 0 | 3 |
| CERS5 | MAP3K13 | 0.726987 | 4 | 0 | 3 |
| CERS5 | TNFRSF10B | 0.726714 | 4 | 0 | 4 |
| CERS5 | DNAH14 | 0.723401 | 4 | 0 | 3 |
| CERS5 | PREB | 0.7154 | 3 | 0 | 3 |
| CERS5 | NPLOC4 | 0.715337 | 3 | 0 | 3 |
| CERS5 | PFN2 | 0.715142 | 5 | 0 | 4 |
| CERS5 | ATM | 0.714093 | 3 | 0 | 3 |
| CERS5 | TMEM187 | 0.713981 | 3 | 0 | 3 |
| CERS5 | AKR1B1 | 0.713808 | 4 | 0 | 4 |
| CERS5 | UBAP2L | 0.712841 | 3 | 0 | 3 |
For details and further investigation, click here