| Full name: ELAV like RNA binding protein 2 | Alias Symbol: HuB|HEL-N1 | ||
| Type: protein-coding gene | Cytoband: 9p21.3 | ||
| Entrez ID: 1993 | HGNC ID: HGNC:3313 | Ensembl Gene: ENSG00000107105 | OMIM ID: 601673 |
| Drug and gene relationship at DGIdb | |||
Expression of ELAVL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ELAVL2 | 1993 | 228260_at | 0.8486 | 0.3063 | |
| GSE20347 | ELAVL2 | 1993 | 208427_s_at | 0.0898 | 0.1493 | |
| GSE23400 | ELAVL2 | 1993 | 208427_s_at | 0.0029 | 0.9388 | |
| GSE26886 | ELAVL2 | 1993 | 228260_at | 0.6305 | 0.1947 | |
| GSE29001 | ELAVL2 | 1993 | 208427_s_at | 0.1394 | 0.5483 | |
| GSE38129 | ELAVL2 | 1993 | 208427_s_at | 0.0077 | 0.9165 | |
| GSE45670 | ELAVL2 | 1993 | 228260_at | 1.5369 | 0.0407 | |
| GSE53622 | ELAVL2 | 1993 | 126872 | 0.7539 | 0.0003 | |
| GSE53624 | ELAVL2 | 1993 | 126872 | 1.0223 | 0.0000 | |
| GSE63941 | ELAVL2 | 1993 | 228260_at | 2.2529 | 0.1604 | |
| GSE77861 | ELAVL2 | 1993 | 208427_s_at | -0.0331 | 0.7649 | |
| TCGA | ELAVL2 | 1993 | RNAseq | 0.1616 | 0.8039 |
Upregulated datasets: 2; Downregulated datasets: 0.
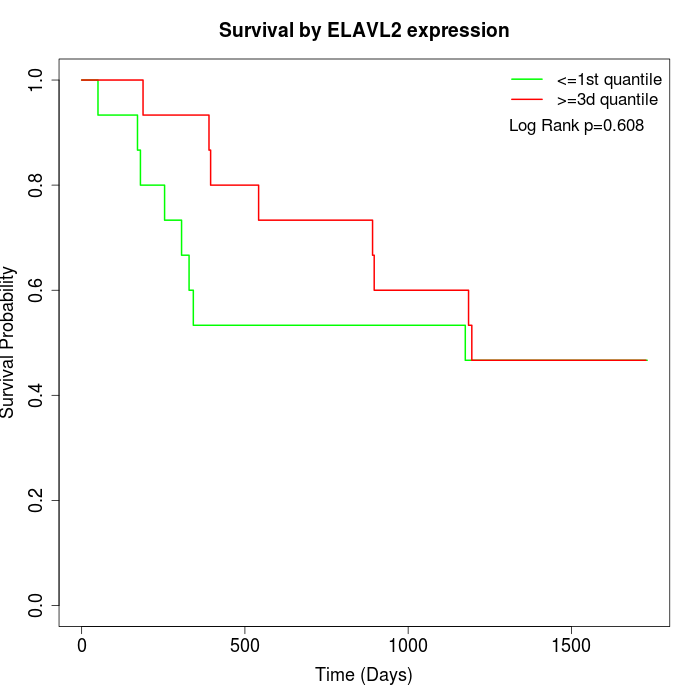
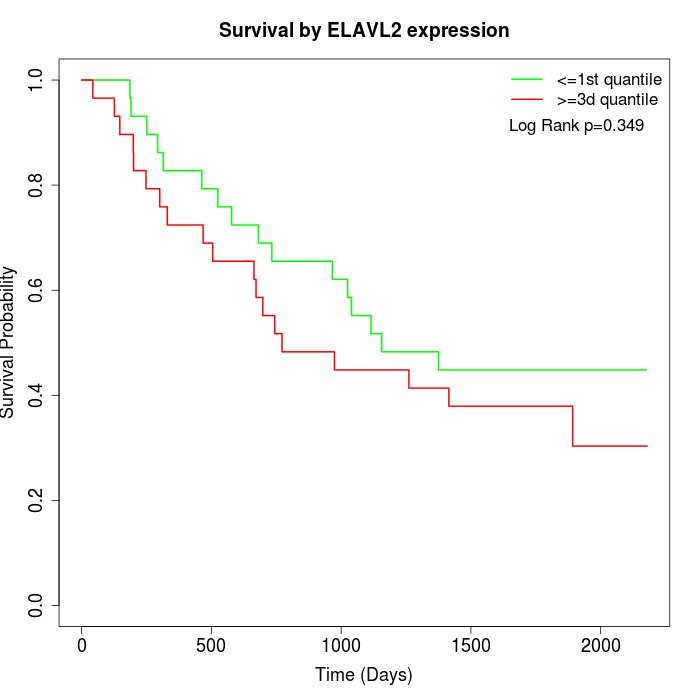
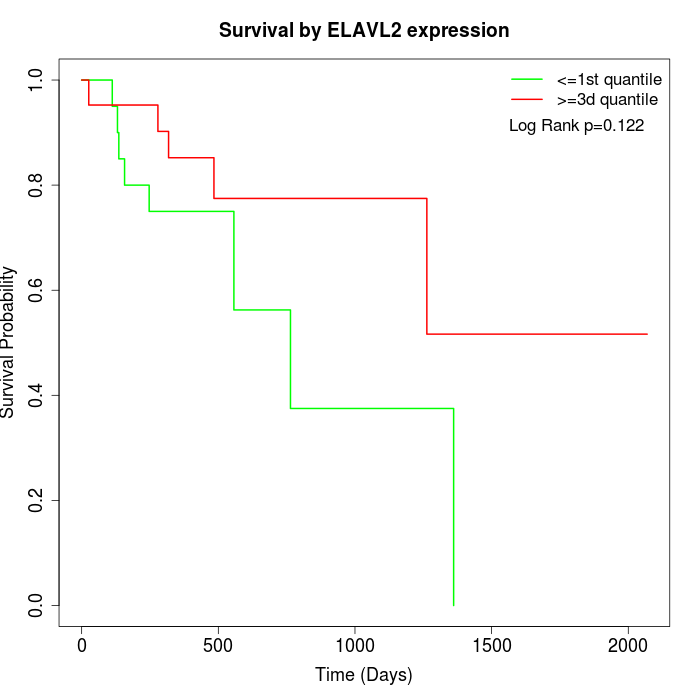
Survival by ELAVL2 expression:
Note: Click image to view full size file.
Copy number change of ELAVL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ELAVL2 | 1993 | 2 | 16 | 12 | |
| GSE20123 | ELAVL2 | 1993 | 2 | 16 | 12 | |
| GSE43470 | ELAVL2 | 1993 | 1 | 13 | 29 | |
| GSE46452 | ELAVL2 | 1993 | 5 | 20 | 34 | |
| GSE47630 | ELAVL2 | 1993 | 1 | 22 | 17 | |
| GSE54993 | ELAVL2 | 1993 | 8 | 0 | 62 | |
| GSE54994 | ELAVL2 | 1993 | 5 | 19 | 29 | |
| GSE60625 | ELAVL2 | 1993 | 0 | 0 | 11 | |
| GSE74703 | ELAVL2 | 1993 | 1 | 10 | 25 | |
| GSE74704 | ELAVL2 | 1993 | 0 | 13 | 7 | |
| TCGA | ELAVL2 | 1993 | 13 | 51 | 32 |
Total number of gains: 38; Total number of losses: 180; Total Number of normals: 270.
Somatic mutations of ELAVL2:
Generating mutation plots.
Highly correlated genes for ELAVL2:
Showing top 20/157 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ELAVL2 | ZFR2 | 0.716974 | 3 | 0 | 3 |
| ELAVL2 | FOLR3 | 0.716587 | 3 | 0 | 3 |
| ELAVL2 | KCNV1 | 0.692644 | 4 | 0 | 4 |
| ELAVL2 | PRRT3-AS1 | 0.670927 | 3 | 0 | 3 |
| ELAVL2 | KIR3DL3 | 0.657536 | 5 | 0 | 4 |
| ELAVL2 | CCR8 | 0.642444 | 4 | 0 | 3 |
| ELAVL2 | FOXN3-AS2 | 0.640201 | 5 | 0 | 5 |
| ELAVL2 | CCR4 | 0.639334 | 3 | 0 | 3 |
| ELAVL2 | RARA | 0.635296 | 3 | 0 | 3 |
| ELAVL2 | MPO | 0.633899 | 3 | 0 | 3 |
| ELAVL2 | SYT12 | 0.627293 | 3 | 0 | 3 |
| ELAVL2 | GPA33 | 0.625553 | 4 | 0 | 3 |
| ELAVL2 | ATG10 | 0.625496 | 4 | 0 | 3 |
| ELAVL2 | CDON | 0.623289 | 3 | 0 | 3 |
| ELAVL2 | FOLH1 | 0.61284 | 4 | 0 | 3 |
| ELAVL2 | SCN11A | 0.608057 | 4 | 0 | 3 |
| ELAVL2 | CBL | 0.605387 | 6 | 0 | 5 |
| ELAVL2 | IL24 | 0.60476 | 4 | 0 | 4 |
| ELAVL2 | DNAJC9 | 0.603805 | 3 | 0 | 3 |
| ELAVL2 | CHST5 | 0.602512 | 3 | 0 | 3 |
For details and further investigation, click here