| Full name: PROP paired-like homeobox 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 5q35.3 | ||
| Entrez ID: 5626 | HGNC ID: HGNC:9455 | Ensembl Gene: ENSG00000175325 | OMIM ID: 601538 |
| Drug and gene relationship at DGIdb | |||
Expression of PROP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PROP1 | 5626 | 211223_at | 0.0072 | 0.9830 | |
| GSE20347 | PROP1 | 5626 | 211223_at | -0.0137 | 0.8974 | |
| GSE23400 | PROP1 | 5626 | 211223_at | -0.1239 | 0.0015 | |
| GSE26886 | PROP1 | 5626 | 211223_at | 0.1594 | 0.1303 | |
| GSE29001 | PROP1 | 5626 | 211223_at | -0.2284 | 0.1165 | |
| GSE38129 | PROP1 | 5626 | 211223_at | -0.1620 | 0.2445 | |
| GSE45670 | PROP1 | 5626 | 211223_at | -0.0717 | 0.5503 | |
| GSE53622 | PROP1 | 5626 | 55135 | -0.4682 | 0.0000 | |
| GSE53624 | PROP1 | 5626 | 55135 | -0.2374 | 0.0026 | |
| GSE63941 | PROP1 | 5626 | 211223_at | 0.0696 | 0.6289 | |
| GSE77861 | PROP1 | 5626 | 211223_at | -0.0855 | 0.6582 | |
| TCGA | PROP1 | 5626 | RNAseq | -0.6469 | 0.5810 |
Upregulated datasets: 0; Downregulated datasets: 0.
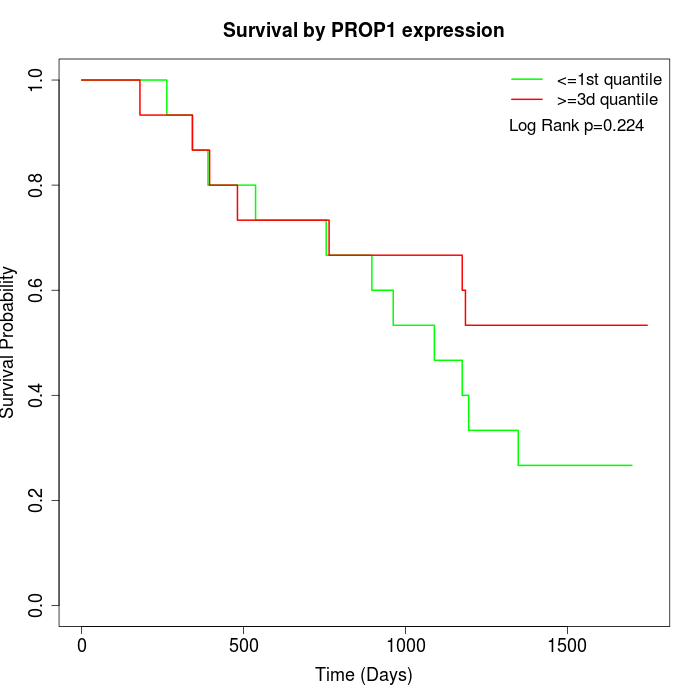
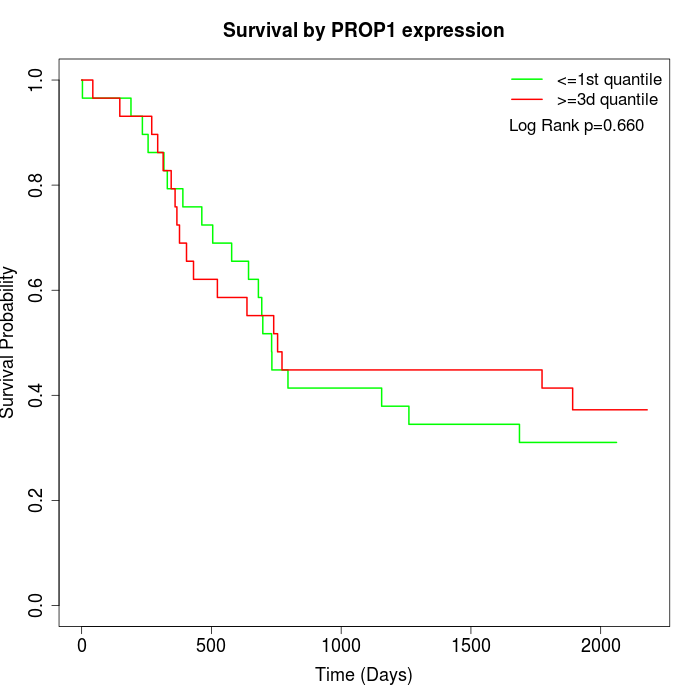
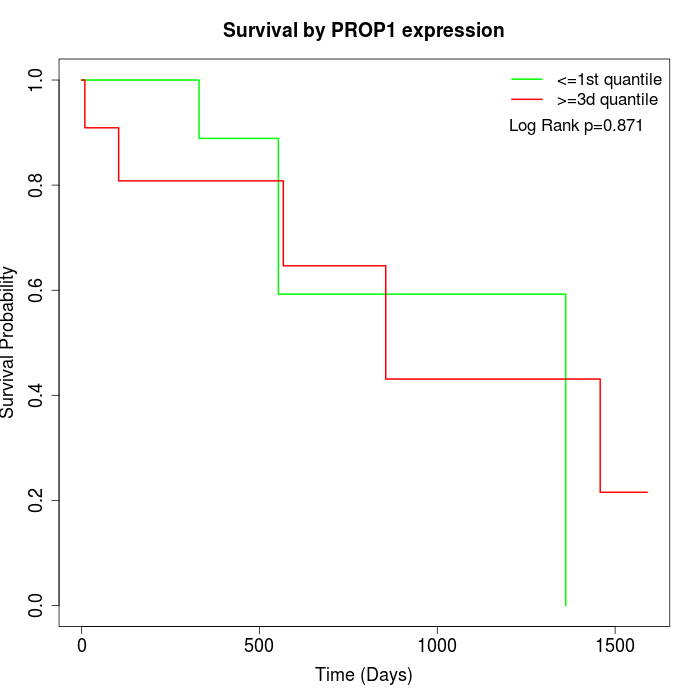
Survival by PROP1 expression:
Note: Click image to view full size file.
Copy number change of PROP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PROP1 | 5626 | 2 | 13 | 15 | |
| GSE20123 | PROP1 | 5626 | 2 | 13 | 15 | |
| GSE43470 | PROP1 | 5626 | 1 | 11 | 31 | |
| GSE46452 | PROP1 | 5626 | 0 | 27 | 32 | |
| GSE47630 | PROP1 | 5626 | 1 | 20 | 19 | |
| GSE54993 | PROP1 | 5626 | 10 | 2 | 58 | |
| GSE54994 | PROP1 | 5626 | 2 | 17 | 34 | |
| GSE60625 | PROP1 | 5626 | 1 | 0 | 10 | |
| GSE74703 | PROP1 | 5626 | 1 | 7 | 28 | |
| GSE74704 | PROP1 | 5626 | 1 | 6 | 13 | |
| TCGA | PROP1 | 5626 | 8 | 34 | 54 |
Total number of gains: 29; Total number of losses: 150; Total Number of normals: 309.
Somatic mutations of PROP1:
Generating mutation plots.
Highly correlated genes for PROP1:
Showing top 20/930 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PROP1 | OR10H3 | 0.734728 | 4 | 0 | 4 |
| PROP1 | SELPLG | 0.7237 | 4 | 0 | 4 |
| PROP1 | RHAG | 0.70127 | 5 | 0 | 5 |
| PROP1 | ELSPBP1 | 0.699407 | 4 | 0 | 4 |
| PROP1 | RAB3IL1 | 0.697353 | 4 | 0 | 4 |
| PROP1 | GDF5 | 0.692877 | 6 | 0 | 5 |
| PROP1 | SPAG11A | 0.691378 | 5 | 0 | 5 |
| PROP1 | MYO1A | 0.690059 | 4 | 0 | 4 |
| PROP1 | PAPPA2 | 0.689069 | 5 | 0 | 5 |
| PROP1 | CCDC71 | 0.687588 | 3 | 0 | 3 |
| PROP1 | BRINP1 | 0.682584 | 3 | 0 | 3 |
| PROP1 | GJB1 | 0.681312 | 6 | 0 | 5 |
| PROP1 | DRD5 | 0.680325 | 6 | 0 | 6 |
| PROP1 | CAMK2B | 0.678579 | 5 | 0 | 5 |
| PROP1 | TAS2R1 | 0.668575 | 4 | 0 | 4 |
| PROP1 | RAI1 | 0.665825 | 4 | 0 | 4 |
| PROP1 | INE1 | 0.664868 | 5 | 0 | 5 |
| PROP1 | XCR1 | 0.664543 | 5 | 0 | 5 |
| PROP1 | IKZF4 | 0.664369 | 3 | 0 | 3 |
| PROP1 | AATK | 0.663404 | 5 | 0 | 5 |
For details and further investigation, click here