| Full name: EYA transcriptional coactivator and phosphatase 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 8q13.3 | ||
| Entrez ID: 2138 | HGNC ID: HGNC:3519 | Ensembl Gene: ENSG00000104313 | OMIM ID: 601653 |
| Drug and gene relationship at DGIdb | |||
Expression of EYA1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EYA1 | 2138 | 214608_s_at | -0.2531 | 0.6838 | |
| GSE20347 | EYA1 | 2138 | 214608_s_at | 0.0841 | 0.5491 | |
| GSE23400 | EYA1 | 2138 | 214608_s_at | -0.0600 | 0.1697 | |
| GSE26886 | EYA1 | 2138 | 214608_s_at | 0.3909 | 0.3749 | |
| GSE29001 | EYA1 | 2138 | 214608_s_at | -0.3359 | 0.2095 | |
| GSE38129 | EYA1 | 2138 | 214608_s_at | -0.0561 | 0.6967 | |
| GSE45670 | EYA1 | 2138 | 214608_s_at | -0.2903 | 0.5178 | |
| GSE53622 | EYA1 | 2138 | 66500 | -0.6248 | 0.0019 | |
| GSE53624 | EYA1 | 2138 | 66500 | -0.3161 | 0.0894 | |
| GSE63941 | EYA1 | 2138 | 214608_s_at | -1.9147 | 0.0073 | |
| GSE77861 | EYA1 | 2138 | 214608_s_at | -0.0566 | 0.6076 | |
| SRP133303 | EYA1 | 2138 | RNAseq | 0.5503 | 0.3079 | |
| TCGA | EYA1 | 2138 | RNAseq | -0.5159 | 0.2645 |
Upregulated datasets: 0; Downregulated datasets: 1.
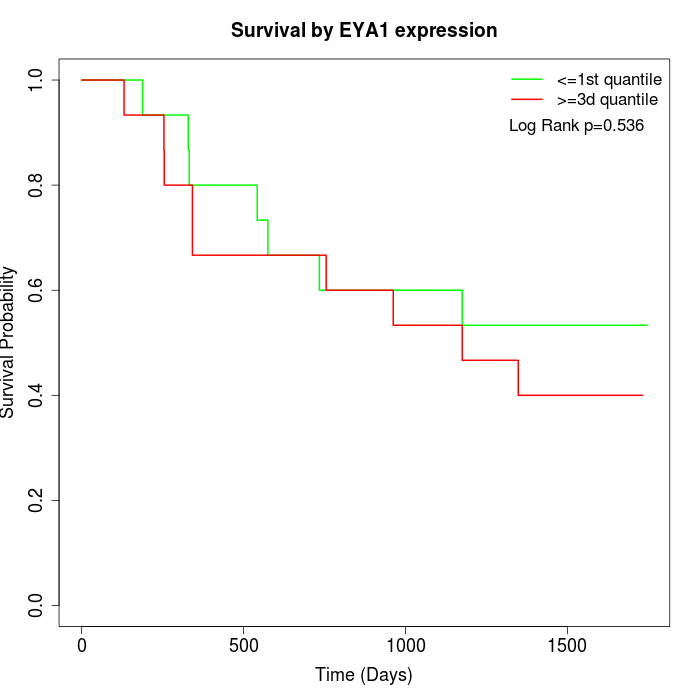
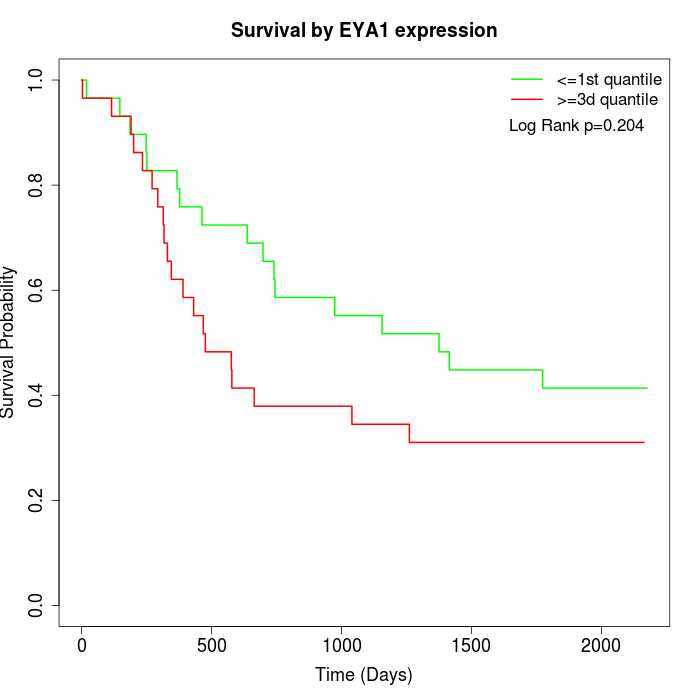
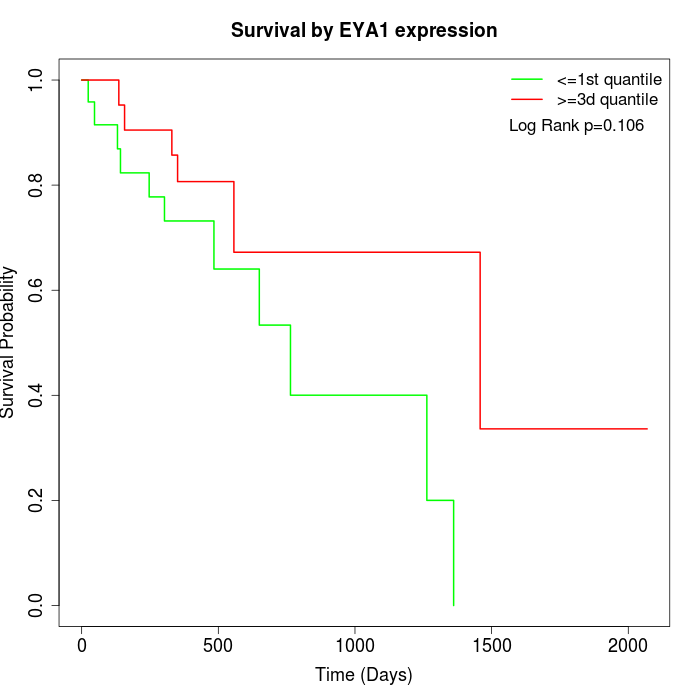
Survival by EYA1 expression:
Note: Click image to view full size file.
Copy number change of EYA1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EYA1 | 2138 | 15 | 1 | 14 | |
| GSE20123 | EYA1 | 2138 | 16 | 1 | 13 | |
| GSE43470 | EYA1 | 2138 | 18 | 1 | 24 | |
| GSE46452 | EYA1 | 2138 | 20 | 2 | 37 | |
| GSE47630 | EYA1 | 2138 | 23 | 0 | 17 | |
| GSE54993 | EYA1 | 2138 | 0 | 19 | 51 | |
| GSE54994 | EYA1 | 2138 | 30 | 0 | 23 | |
| GSE60625 | EYA1 | 2138 | 0 | 4 | 7 | |
| GSE74703 | EYA1 | 2138 | 15 | 1 | 20 | |
| GSE74704 | EYA1 | 2138 | 11 | 0 | 9 | |
| TCGA | EYA1 | 2138 | 51 | 2 | 43 |
Total number of gains: 199; Total number of losses: 31; Total Number of normals: 258.
Somatic mutations of EYA1:
Generating mutation plots.
Highly correlated genes for EYA1:
Showing top 20/61 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EYA1 | CP | 0.771375 | 3 | 0 | 3 |
| EYA1 | NR3C2 | 0.675565 | 3 | 0 | 3 |
| EYA1 | TMEM170B | 0.670436 | 3 | 0 | 3 |
| EYA1 | RERG | 0.668603 | 3 | 0 | 3 |
| EYA1 | WFDC2 | 0.648986 | 4 | 0 | 3 |
| EYA1 | ANO3 | 0.646315 | 3 | 0 | 3 |
| EYA1 | PROM1 | 0.64289 | 3 | 0 | 3 |
| EYA1 | LRCH2 | 0.639898 | 4 | 0 | 4 |
| EYA1 | LAMA2 | 0.638603 | 3 | 0 | 3 |
| EYA1 | MLPH | 0.635819 | 4 | 0 | 3 |
| EYA1 | GRIA2 | 0.631423 | 3 | 0 | 3 |
| EYA1 | RAB37 | 0.625981 | 3 | 0 | 3 |
| EYA1 | SLAIN1 | 0.625279 | 4 | 0 | 3 |
| EYA1 | KCNT2 | 0.609175 | 5 | 0 | 5 |
| EYA1 | KIAA1107 | 0.606419 | 4 | 0 | 3 |
| EYA1 | LPIN2 | 0.597098 | 3 | 0 | 3 |
| EYA1 | MAP9 | 0.594504 | 3 | 0 | 3 |
| EYA1 | MICAL1 | 0.592788 | 3 | 0 | 3 |
| EYA1 | RUNX1T1 | 0.591689 | 4 | 0 | 3 |
| EYA1 | SCN7A | 0.590245 | 3 | 0 | 3 |
For details and further investigation, click here