| Full name: potassium sodium-activated channel subfamily T member 2 | Alias Symbol: KCa4.2|SLICK|SLO2.1 | ||
| Type: protein-coding gene | Cytoband: 1q31.3 | ||
| Entrez ID: 343450 | HGNC ID: HGNC:18866 | Ensembl Gene: ENSG00000162687 | OMIM ID: 610044 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNT2 | 343450 | 244455_at | -1.6881 | 0.1434 | |
| GSE26886 | KCNT2 | 343450 | 244455_at | 0.8324 | 0.0720 | |
| GSE45670 | KCNT2 | 343450 | 244455_at | -2.4118 | 0.0000 | |
| GSE53622 | KCNT2 | 343450 | 12586 | -1.4988 | 0.0000 | |
| GSE53624 | KCNT2 | 343450 | 48042 | -1.5744 | 0.0000 | |
| GSE63941 | KCNT2 | 343450 | 244455_at | -2.8758 | 0.0000 | |
| GSE77861 | KCNT2 | 343450 | 234103_at | 0.0038 | 0.9673 | |
| GSE97050 | KCNT2 | 343450 | A_33_P3892608 | -0.5146 | 0.3562 | |
| SRP007169 | KCNT2 | 343450 | RNAseq | 1.9139 | 0.0339 | |
| SRP133303 | KCNT2 | 343450 | RNAseq | -1.5873 | 0.0000 | |
| SRP159526 | KCNT2 | 343450 | RNAseq | -0.0538 | 0.9171 | |
| SRP219564 | KCNT2 | 343450 | RNAseq | 1.0534 | 0.3322 | |
| TCGA | KCNT2 | 343450 | RNAseq | -1.1361 | 0.0035 |
Upregulated datasets: 1; Downregulated datasets: 6.
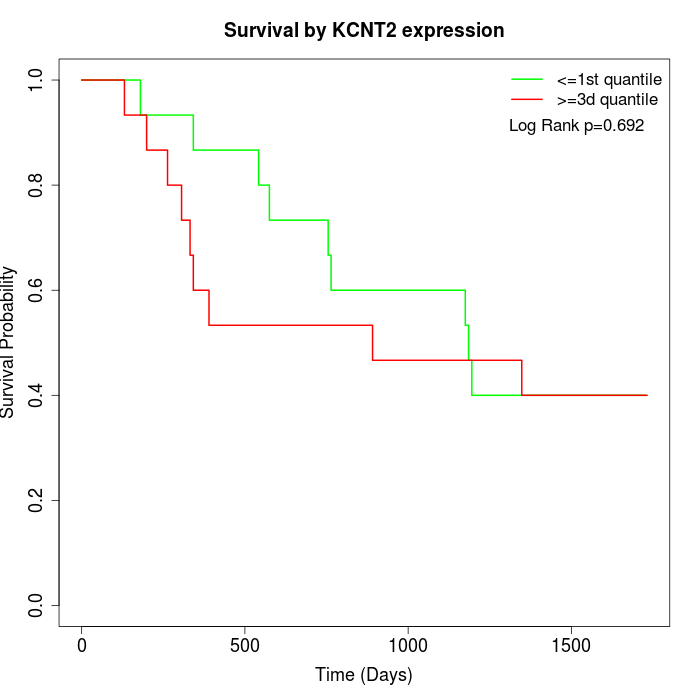
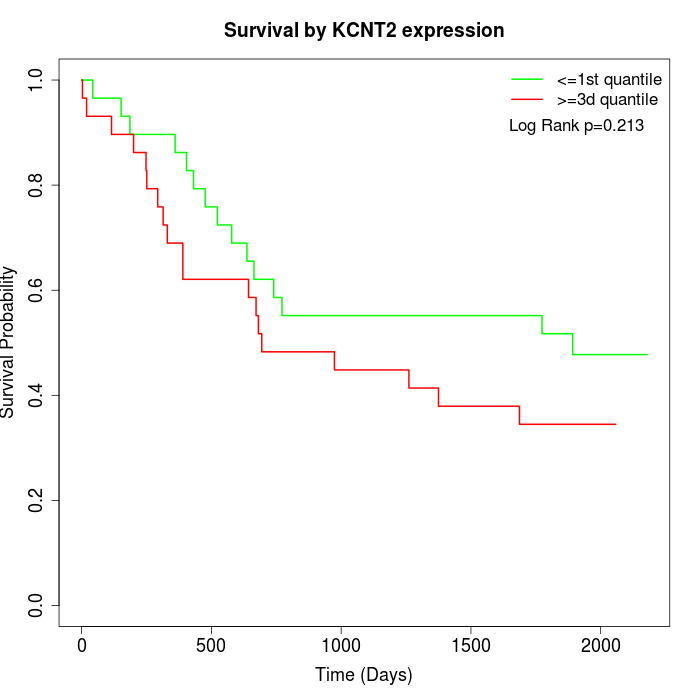
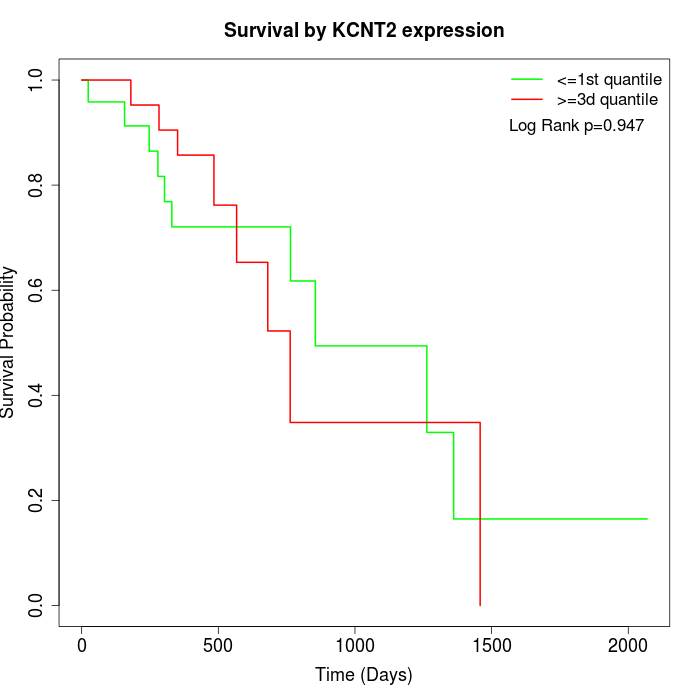
Survival by KCNT2 expression:
Note: Click image to view full size file.
Copy number change of KCNT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNT2 | 343450 | 6 | 1 | 23 | |
| GSE20123 | KCNT2 | 343450 | 6 | 1 | 23 | |
| GSE43470 | KCNT2 | 343450 | 5 | 1 | 37 | |
| GSE46452 | KCNT2 | 343450 | 3 | 1 | 55 | |
| GSE47630 | KCNT2 | 343450 | 14 | 0 | 26 | |
| GSE54993 | KCNT2 | 343450 | 0 | 6 | 64 | |
| GSE54994 | KCNT2 | 343450 | 16 | 0 | 37 | |
| GSE60625 | KCNT2 | 343450 | 0 | 0 | 11 | |
| GSE74703 | KCNT2 | 343450 | 5 | 1 | 30 | |
| GSE74704 | KCNT2 | 343450 | 2 | 1 | 17 | |
| TCGA | KCNT2 | 343450 | 41 | 6 | 49 |
Total number of gains: 98; Total number of losses: 18; Total Number of normals: 372.
Somatic mutations of KCNT2:
Generating mutation plots.
Highly correlated genes for KCNT2:
Showing top 20/1179 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNT2 | FAM13C | 0.850124 | 4 | 0 | 4 |
| KCNT2 | JAM2 | 0.837207 | 6 | 0 | 6 |
| KCNT2 | GYPC | 0.834379 | 6 | 0 | 6 |
| KCNT2 | BHMT2 | 0.831856 | 6 | 0 | 6 |
| KCNT2 | CASQ2 | 0.82903 | 5 | 0 | 5 |
| KCNT2 | PDE5A | 0.820332 | 6 | 0 | 6 |
| KCNT2 | MAGI2-AS3 | 0.816579 | 5 | 0 | 5 |
| KCNT2 | RSPO3 | 0.814337 | 7 | 0 | 7 |
| KCNT2 | MFAP4 | 0.809902 | 6 | 0 | 6 |
| KCNT2 | ASPA | 0.809083 | 6 | 0 | 6 |
| KCNT2 | PGM5 | 0.806839 | 5 | 0 | 5 |
| KCNT2 | FIBIN | 0.806055 | 4 | 0 | 4 |
| KCNT2 | RNF150 | 0.805798 | 7 | 0 | 7 |
| KCNT2 | FAT4 | 0.805729 | 3 | 0 | 3 |
| KCNT2 | AOC3 | 0.805661 | 6 | 0 | 6 |
| KCNT2 | RCAN2 | 0.803317 | 6 | 0 | 6 |
| KCNT2 | CRBN | 0.802534 | 3 | 0 | 3 |
| KCNT2 | SMOC2 | 0.802527 | 5 | 0 | 5 |
| KCNT2 | MAN1C1 | 0.800103 | 6 | 0 | 6 |
| KCNT2 | CILP | 0.799027 | 5 | 0 | 5 |
For details and further investigation, click here