| Full name: fatty acid desaturase 2 | Alias Symbol: FADSD6|D6D|TU13|DES6|SLL0262 | ||
| Type: protein-coding gene | Cytoband: 11q12.2 | ||
| Entrez ID: 9415 | HGNC ID: HGNC:3575 | Ensembl Gene: ENSG00000134824 | OMIM ID: 606149 |
| Drug and gene relationship at DGIdb | |||
FADS2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa03320 | PPAR signaling pathway |
Expression of FADS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FADS2 | 9415 | 202218_s_at | 0.8213 | 0.1498 | |
| GSE20347 | FADS2 | 9415 | 202218_s_at | 0.6608 | 0.0000 | |
| GSE23400 | FADS2 | 9415 | 202218_s_at | 0.4094 | 0.0000 | |
| GSE26886 | FADS2 | 9415 | 202218_s_at | 0.4832 | 0.0001 | |
| GSE29001 | FADS2 | 9415 | 202218_s_at | 0.2158 | 0.3672 | |
| GSE38129 | FADS2 | 9415 | 202218_s_at | 0.5399 | 0.0004 | |
| GSE45670 | FADS2 | 9415 | 202218_s_at | 0.1361 | 0.3511 | |
| GSE53622 | FADS2 | 9415 | 50606 | 2.1539 | 0.0000 | |
| GSE53624 | FADS2 | 9415 | 50606 | 2.5684 | 0.0000 | |
| GSE63941 | FADS2 | 9415 | 202218_s_at | -0.2586 | 0.5708 | |
| GSE77861 | FADS2 | 9415 | 202218_s_at | 0.4549 | 0.0385 | |
| GSE97050 | FADS2 | 9415 | A_33_P3360326 | 0.0754 | 0.7920 | |
| SRP007169 | FADS2 | 9415 | RNAseq | 5.6246 | 0.0000 | |
| SRP008496 | FADS2 | 9415 | RNAseq | 5.0773 | 0.0000 | |
| SRP064894 | FADS2 | 9415 | RNAseq | 2.3459 | 0.0000 | |
| SRP133303 | FADS2 | 9415 | RNAseq | 2.1447 | 0.0000 | |
| SRP159526 | FADS2 | 9415 | RNAseq | 3.8906 | 0.0000 | |
| SRP193095 | FADS2 | 9415 | RNAseq | 2.8533 | 0.0000 | |
| SRP219564 | FADS2 | 9415 | RNAseq | 1.1743 | 0.2055 | |
| TCGA | FADS2 | 9415 | RNAseq | 0.4883 | 0.0006 |
Upregulated datasets: 8; Downregulated datasets: 0.
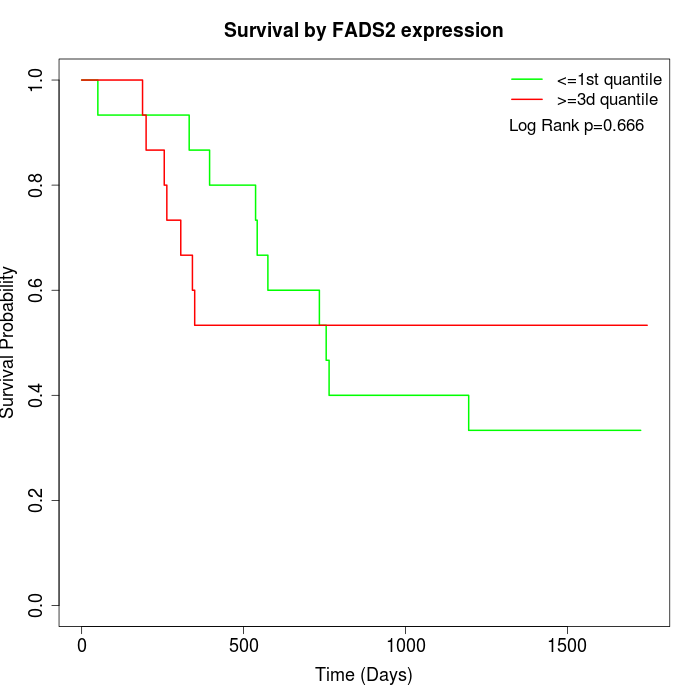
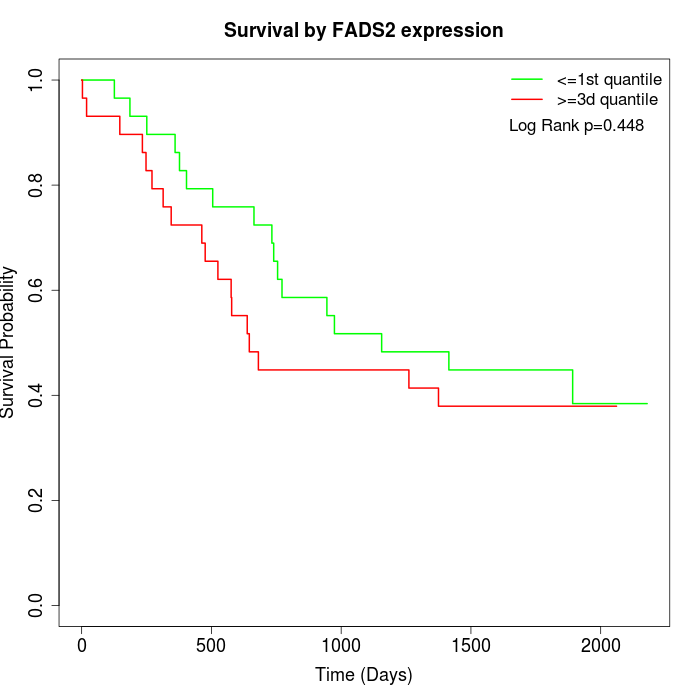
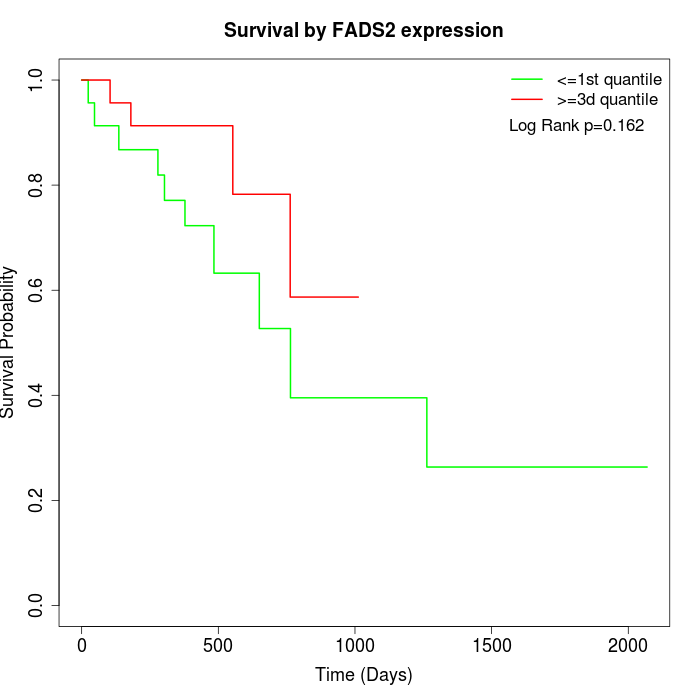
Survival by FADS2 expression:
Note: Click image to view full size file.
Copy number change of FADS2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FADS2 | 9415 | 5 | 5 | 20 | |
| GSE20123 | FADS2 | 9415 | 5 | 5 | 20 | |
| GSE43470 | FADS2 | 9415 | 2 | 6 | 35 | |
| GSE46452 | FADS2 | 9415 | 8 | 4 | 47 | |
| GSE47630 | FADS2 | 9415 | 3 | 7 | 30 | |
| GSE54993 | FADS2 | 9415 | 3 | 0 | 67 | |
| GSE54994 | FADS2 | 9415 | 4 | 8 | 41 | |
| GSE60625 | FADS2 | 9415 | 0 | 3 | 8 | |
| GSE74703 | FADS2 | 9415 | 2 | 3 | 31 | |
| GSE74704 | FADS2 | 9415 | 4 | 3 | 13 | |
| TCGA | FADS2 | 9415 | 14 | 10 | 72 |
Total number of gains: 50; Total number of losses: 54; Total Number of normals: 384.
Somatic mutations of FADS2:
Generating mutation plots.
Highly correlated genes for FADS2:
Showing top 20/1148 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FADS2 | ZNF114 | 0.747202 | 3 | 0 | 3 |
| FADS2 | COL12A1 | 0.746207 | 5 | 0 | 5 |
| FADS2 | WDR54 | 0.740043 | 4 | 0 | 4 |
| FADS2 | FADS1 | 0.715853 | 4 | 0 | 4 |
| FADS2 | CTHRC1 | 0.706226 | 5 | 0 | 5 |
| FADS2 | KRTCAP2 | 0.697933 | 3 | 0 | 3 |
| FADS2 | FADS3 | 0.696849 | 8 | 0 | 8 |
| FADS2 | FMNL2 | 0.690685 | 5 | 0 | 5 |
| FADS2 | TMEM185B | 0.688578 | 6 | 0 | 6 |
| FADS2 | CMTM3 | 0.68758 | 4 | 0 | 4 |
| FADS2 | CORO7 | 0.68751 | 3 | 0 | 3 |
| FADS2 | NAT14 | 0.684739 | 5 | 0 | 4 |
| FADS2 | GLIS2 | 0.682235 | 5 | 0 | 5 |
| FADS2 | RAB34 | 0.681064 | 3 | 0 | 3 |
| FADS2 | USF2 | 0.678368 | 3 | 0 | 3 |
| FADS2 | DOHH | 0.67832 | 3 | 0 | 3 |
| FADS2 | B3GALT6 | 0.675049 | 4 | 0 | 4 |
| FADS2 | MIER2 | 0.669537 | 3 | 0 | 3 |
| FADS2 | HDAC7 | 0.669443 | 3 | 0 | 3 |
| FADS2 | IKBIP | 0.668957 | 4 | 0 | 4 |
For details and further investigation, click here