| Full name: gamma-aminobutyric acid type B receptor subunit 2 | Alias Symbol: HG20|GABABR2|GPRC3B | ||
| Type: protein-coding gene | Cytoband: 9q22.33 | ||
| Entrez ID: 9568 | HGNC ID: HGNC:4507 | Ensembl Gene: ENSG00000136928 | OMIM ID: 607340 |
| Drug and gene relationship at DGIdb | |||
GABBR2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04024 | cAMP signaling pathway | |
| hsa04727 | GABAergic synapse | |
| hsa04742 | Taste transduction | |
| hsa04915 | Estrogen signaling pathway | |
| hsa05032 | Morphine addiction |
Expression of GABBR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GABBR2 | 9568 | 209991_x_at | -0.0790 | 0.7739 | |
| GSE20347 | GABBR2 | 9568 | 209991_x_at | -0.0776 | 0.3155 | |
| GSE23400 | GABBR2 | 9568 | 211679_x_at | -0.1110 | 0.0143 | |
| GSE26886 | GABBR2 | 9568 | 211679_x_at | 0.2905 | 0.0201 | |
| GSE29001 | GABBR2 | 9568 | 211679_x_at | 0.0068 | 0.9688 | |
| GSE38129 | GABBR2 | 9568 | 209991_x_at | -0.0996 | 0.2366 | |
| GSE45670 | GABBR2 | 9568 | 211679_x_at | 0.2048 | 0.0215 | |
| GSE53622 | GABBR2 | 9568 | 57352 | 0.7949 | 0.0009 | |
| GSE53624 | GABBR2 | 9568 | 57352 | 1.3674 | 0.0000 | |
| GSE63941 | GABBR2 | 9568 | 211679_x_at | -0.0449 | 0.9310 | |
| GSE77861 | GABBR2 | 9568 | 211679_x_at | -0.1708 | 0.0514 | |
| SRP133303 | GABBR2 | 9568 | RNAseq | 0.2788 | 0.2635 | |
| TCGA | GABBR2 | 9568 | RNAseq | -0.4205 | 0.4886 |
Upregulated datasets: 1; Downregulated datasets: 0.
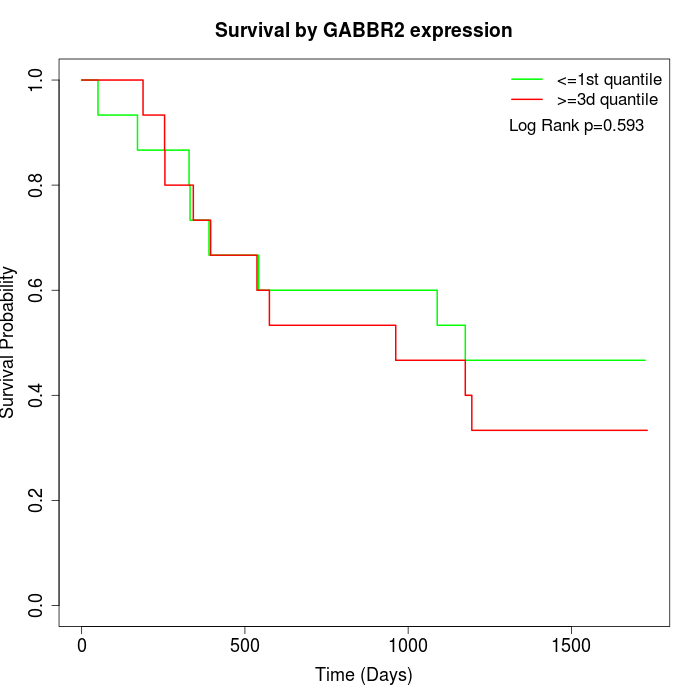
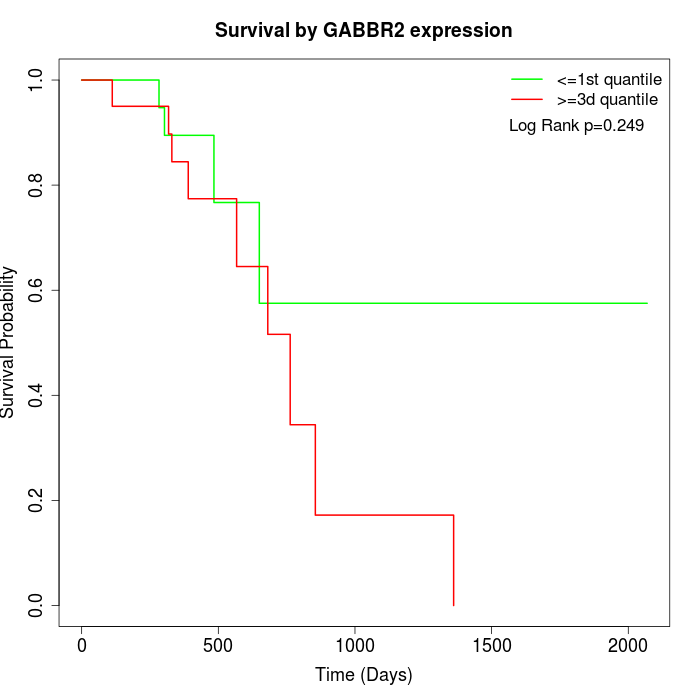
Survival by GABBR2 expression:
Note: Click image to view full size file.
Copy number change of GABBR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GABBR2 | 9568 | 5 | 9 | 16 | |
| GSE20123 | GABBR2 | 9568 | 5 | 8 | 17 | |
| GSE43470 | GABBR2 | 9568 | 6 | 3 | 34 | |
| GSE46452 | GABBR2 | 9568 | 6 | 14 | 39 | |
| GSE47630 | GABBR2 | 9568 | 1 | 17 | 22 | |
| GSE54993 | GABBR2 | 9568 | 3 | 3 | 64 | |
| GSE54994 | GABBR2 | 9568 | 8 | 11 | 34 | |
| GSE60625 | GABBR2 | 9568 | 0 | 0 | 11 | |
| GSE74703 | GABBR2 | 9568 | 5 | 3 | 28 | |
| GSE74704 | GABBR2 | 9568 | 3 | 5 | 12 | |
| TCGA | GABBR2 | 9568 | 26 | 22 | 48 |
Total number of gains: 68; Total number of losses: 95; Total Number of normals: 325.
Somatic mutations of GABBR2:
Generating mutation plots.
Highly correlated genes for GABBR2:
Showing top 20/53 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GABBR2 | DOCK10 | 0.652687 | 3 | 0 | 3 |
| GABBR2 | C12orf42 | 0.628325 | 3 | 0 | 3 |
| GABBR2 | COL4A4 | 0.617734 | 3 | 0 | 3 |
| GABBR2 | RNASEL | 0.589793 | 3 | 0 | 3 |
| GABBR2 | CCDC63 | 0.584527 | 3 | 0 | 3 |
| GABBR2 | MED31 | 0.578352 | 3 | 0 | 3 |
| GABBR2 | PSG1 | 0.577047 | 4 | 0 | 4 |
| GABBR2 | GPLD1 | 0.568499 | 5 | 0 | 4 |
| GABBR2 | GPR17 | 0.567278 | 3 | 0 | 3 |
| GABBR2 | CCL27 | 0.561579 | 4 | 0 | 3 |
| GABBR2 | CER1 | 0.561351 | 4 | 0 | 4 |
| GABBR2 | PIK3R5 | 0.55932 | 4 | 0 | 3 |
| GABBR2 | SERPINA4 | 0.556622 | 4 | 0 | 3 |
| GABBR2 | ABCC8 | 0.554615 | 6 | 0 | 4 |
| GABBR2 | MYL1 | 0.554545 | 4 | 0 | 3 |
| GABBR2 | CBLN1 | 0.550184 | 3 | 0 | 3 |
| GABBR2 | ZNF280A | 0.548858 | 5 | 0 | 3 |
| GABBR2 | CHIT1 | 0.54349 | 3 | 0 | 3 |
| GABBR2 | OR7C1 | 0.542072 | 4 | 0 | 3 |
| GABBR2 | S100G | 0.539351 | 6 | 0 | 3 |
For details and further investigation, click here