| Full name: growth arrest and DNA damage inducible gamma | Alias Symbol: DDIT2|GADD45gamma|GRP17|CR6 | ||
| Type: protein-coding gene | Cytoband: 9q22.2 | ||
| Entrez ID: 10912 | HGNC ID: HGNC:4097 | Ensembl Gene: ENSG00000130222 | OMIM ID: 604949 |
| Drug and gene relationship at DGIdb | |||
GADD45G involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04068 | FoxO signaling pathway | |
| hsa04110 | Cell cycle | |
| hsa04115 | p53 signaling pathway | |
| hsa04210 | Apoptosis |
Expression of GADD45G:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GADD45G | 10912 | 204121_at | -1.1381 | 0.4801 | |
| GSE20347 | GADD45G | 10912 | 204121_at | -0.2743 | 0.1063 | |
| GSE23400 | GADD45G | 10912 | 204121_at | -0.2082 | 0.0007 | |
| GSE26886 | GADD45G | 10912 | 204121_at | 0.3476 | 0.0845 | |
| GSE29001 | GADD45G | 10912 | 204121_at | -0.0646 | 0.8737 | |
| GSE38129 | GADD45G | 10912 | 204121_at | -0.6375 | 0.0168 | |
| GSE45670 | GADD45G | 10912 | 204121_at | -0.5921 | 0.0112 | |
| GSE53622 | GADD45G | 10912 | 78337 | -0.6956 | 0.0000 | |
| GSE53624 | GADD45G | 10912 | 78337 | -0.7224 | 0.0000 | |
| GSE63941 | GADD45G | 10912 | 204121_at | -0.0842 | 0.8827 | |
| GSE77861 | GADD45G | 10912 | 204121_at | -0.0375 | 0.8856 | |
| GSE97050 | GADD45G | 10912 | A_33_P3252394 | -1.1341 | 0.0981 | |
| SRP064894 | GADD45G | 10912 | RNAseq | -1.4136 | 0.0036 | |
| SRP133303 | GADD45G | 10912 | RNAseq | -0.4560 | 0.2813 | |
| SRP159526 | GADD45G | 10912 | RNAseq | -0.9535 | 0.1406 | |
| SRP219564 | GADD45G | 10912 | RNAseq | -1.3002 | 0.2647 | |
| TCGA | GADD45G | 10912 | RNAseq | -0.6563 | 0.0010 |
Upregulated datasets: 0; Downregulated datasets: 1.
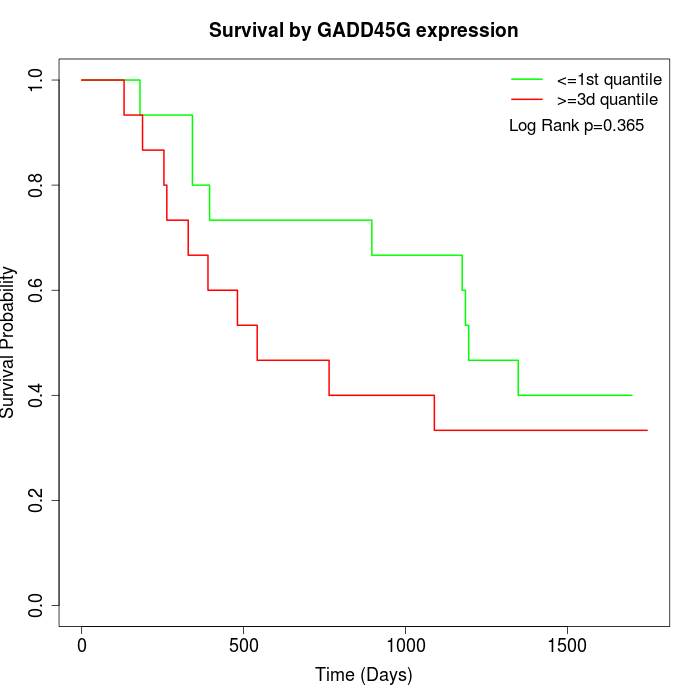
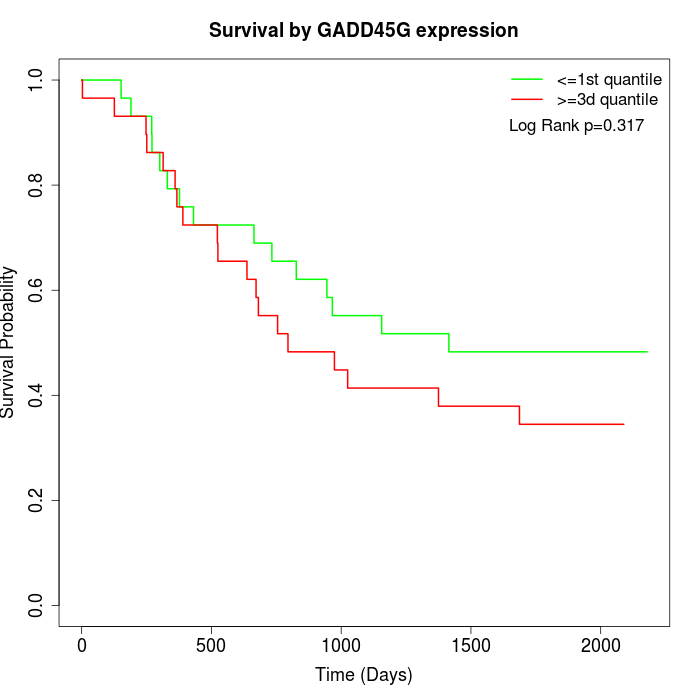
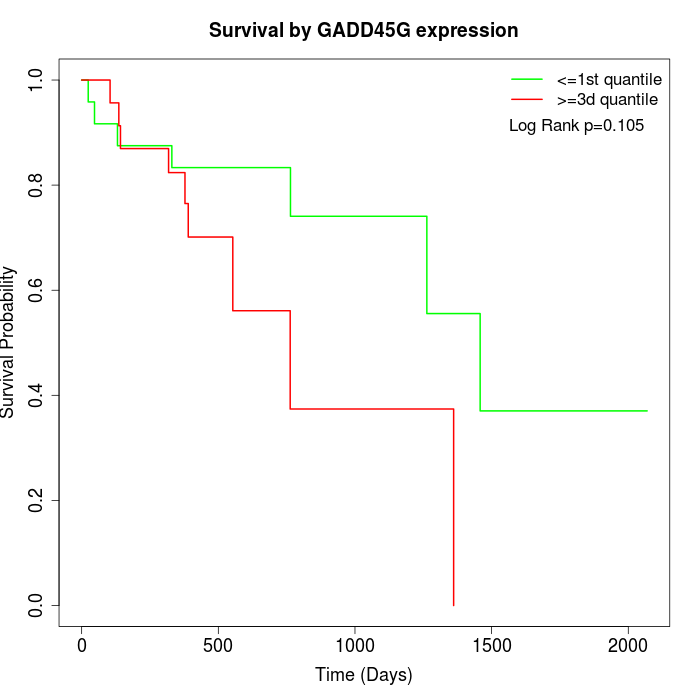
Survival by GADD45G expression:
Note: Click image to view full size file.
Copy number change of GADD45G:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GADD45G | 10912 | 5 | 10 | 15 | |
| GSE20123 | GADD45G | 10912 | 5 | 10 | 15 | |
| GSE43470 | GADD45G | 10912 | 6 | 3 | 34 | |
| GSE46452 | GADD45G | 10912 | 6 | 14 | 39 | |
| GSE47630 | GADD45G | 10912 | 1 | 18 | 21 | |
| GSE54993 | GADD45G | 10912 | 4 | 2 | 64 | |
| GSE54994 | GADD45G | 10912 | 7 | 11 | 35 | |
| GSE60625 | GADD45G | 10912 | 0 | 0 | 11 | |
| GSE74703 | GADD45G | 10912 | 5 | 3 | 28 | |
| GSE74704 | GADD45G | 10912 | 3 | 7 | 10 | |
| TCGA | GADD45G | 10912 | 21 | 24 | 51 |
Total number of gains: 63; Total number of losses: 102; Total Number of normals: 323.
Somatic mutations of GADD45G:
Generating mutation plots.
Highly correlated genes for GADD45G:
Showing top 20/530 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GADD45G | DDX42 | 0.763283 | 3 | 0 | 3 |
| GADD45G | PDK2 | 0.727825 | 3 | 0 | 3 |
| GADD45G | SMG6 | 0.708923 | 4 | 0 | 4 |
| GADD45G | NFIC | 0.70255 | 6 | 0 | 5 |
| GADD45G | TTLL11 | 0.699606 | 3 | 0 | 3 |
| GADD45G | POLR1D | 0.696011 | 3 | 0 | 3 |
| GADD45G | GKAP1 | 0.690299 | 3 | 0 | 3 |
| GADD45G | APLP2 | 0.685448 | 3 | 0 | 3 |
| GADD45G | ZNF383 | 0.671999 | 3 | 0 | 3 |
| GADD45G | PDCD4 | 0.667516 | 4 | 0 | 4 |
| GADD45G | CLEC3B | 0.664486 | 3 | 0 | 3 |
| GADD45G | COX7C | 0.658492 | 4 | 0 | 3 |
| GADD45G | FZD7 | 0.656181 | 4 | 0 | 3 |
| GADD45G | ZBTB16 | 0.649828 | 7 | 0 | 7 |
| GADD45G | CYP21A2 | 0.649177 | 3 | 0 | 3 |
| GADD45G | NR1H2 | 0.645913 | 4 | 0 | 4 |
| GADD45G | CCDC92 | 0.643341 | 5 | 0 | 4 |
| GADD45G | CLK4 | 0.640359 | 3 | 0 | 3 |
| GADD45G | RNF115 | 0.639681 | 6 | 0 | 4 |
| GADD45G | RHOB | 0.638604 | 7 | 0 | 6 |
For details and further investigation, click here