| Full name: glycine amidinotransferase | Alias Symbol: AGAT | ||
| Type: protein-coding gene | Cytoband: 15q21.1 | ||
| Entrez ID: 2628 | HGNC ID: HGNC:4175 | Ensembl Gene: ENSG00000171766 | OMIM ID: 602360 |
| Drug and gene relationship at DGIdb | |||
Expression of GATM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GATM | 2628 | 203178_at | -1.4622 | 0.1500 | |
| GSE20347 | GATM | 2628 | 203178_at | -1.4071 | 0.0064 | |
| GSE23400 | GATM | 2628 | 216733_s_at | -0.8928 | 0.0000 | |
| GSE26886 | GATM | 2628 | 203178_at | -1.2038 | 0.0099 | |
| GSE29001 | GATM | 2628 | 216733_s_at | -0.7991 | 0.0055 | |
| GSE38129 | GATM | 2628 | 203178_at | -1.2924 | 0.0002 | |
| GSE45670 | GATM | 2628 | 203178_at | -1.0430 | 0.0484 | |
| GSE53622 | GATM | 2628 | 130788 | -1.9450 | 0.0000 | |
| GSE53624 | GATM | 2628 | 130788 | -2.2458 | 0.0000 | |
| GSE63941 | GATM | 2628 | 216733_s_at | 2.9284 | 0.1400 | |
| GSE77861 | GATM | 2628 | 216733_s_at | -1.6057 | 0.0120 | |
| GSE97050 | GATM | 2628 | A_23_P129064 | -0.5680 | 0.1297 | |
| SRP007169 | GATM | 2628 | RNAseq | -1.1847 | 0.0397 | |
| SRP008496 | GATM | 2628 | RNAseq | -1.6737 | 0.0000 | |
| SRP064894 | GATM | 2628 | RNAseq | -1.7498 | 0.0000 | |
| SRP133303 | GATM | 2628 | RNAseq | -0.9467 | 0.0004 | |
| SRP159526 | GATM | 2628 | RNAseq | -0.9360 | 0.0162 | |
| SRP193095 | GATM | 2628 | RNAseq | -0.9457 | 0.0038 | |
| SRP219564 | GATM | 2628 | RNAseq | -1.4882 | 0.0022 | |
| TCGA | GATM | 2628 | RNAseq | -0.7061 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 11.
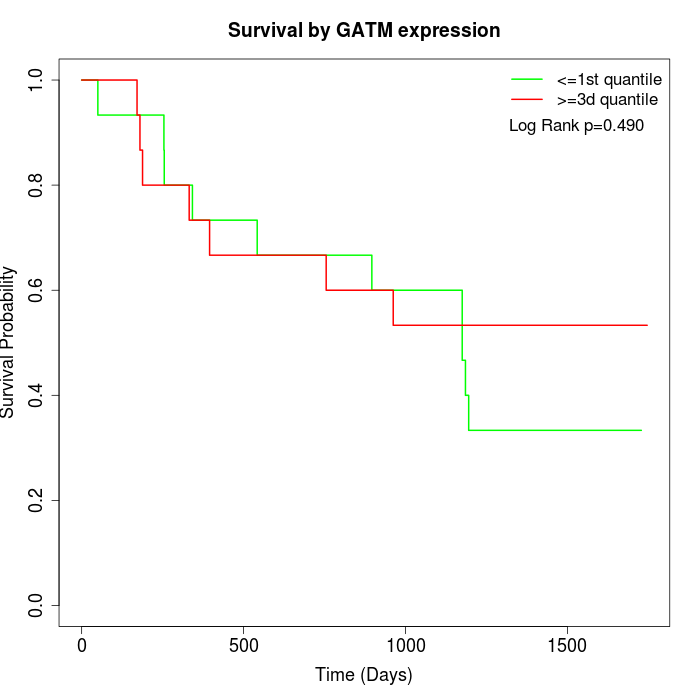
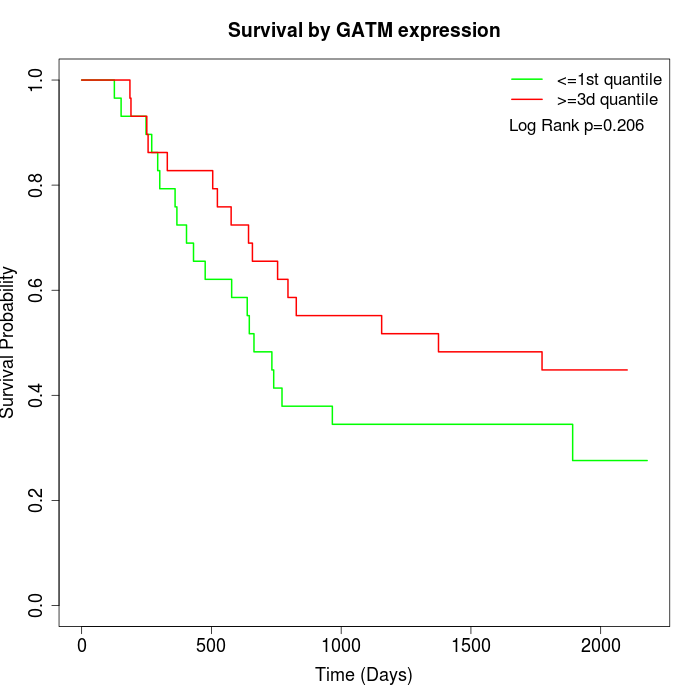
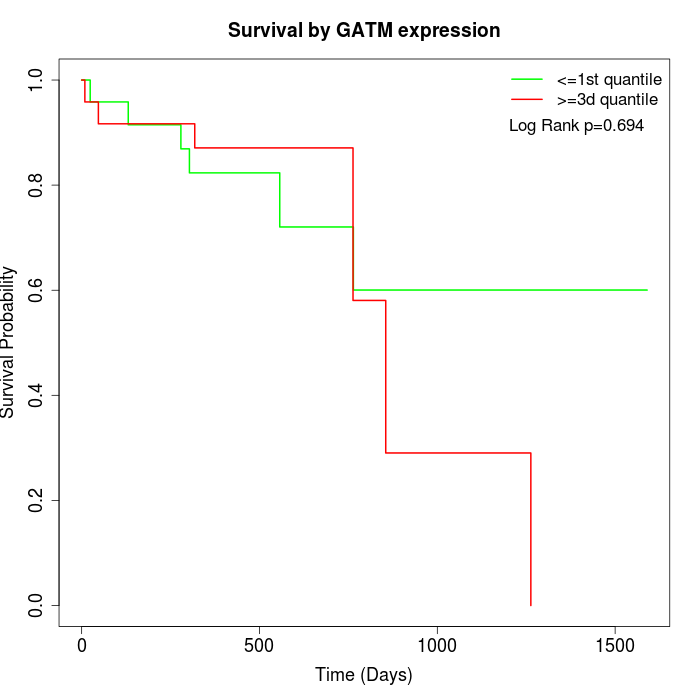
Survival by GATM expression:
Note: Click image to view full size file.
Copy number change of GATM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GATM | 2628 | 4 | 5 | 21 | |
| GSE20123 | GATM | 2628 | 4 | 5 | 21 | |
| GSE43470 | GATM | 2628 | 4 | 4 | 35 | |
| GSE46452 | GATM | 2628 | 3 | 7 | 49 | |
| GSE47630 | GATM | 2628 | 8 | 10 | 22 | |
| GSE54993 | GATM | 2628 | 4 | 5 | 61 | |
| GSE54994 | GATM | 2628 | 5 | 7 | 41 | |
| GSE60625 | GATM | 2628 | 4 | 0 | 7 | |
| GSE74703 | GATM | 2628 | 4 | 3 | 29 | |
| GSE74704 | GATM | 2628 | 2 | 3 | 15 | |
| TCGA | GATM | 2628 | 11 | 16 | 69 |
Total number of gains: 53; Total number of losses: 65; Total Number of normals: 370.
Somatic mutations of GATM:
Generating mutation plots.
Highly correlated genes for GATM:
Showing top 20/1215 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GATM | SVIP | 0.750488 | 6 | 0 | 6 |
| GATM | CA13 | 0.749681 | 3 | 0 | 3 |
| GATM | GBP6 | 0.743465 | 3 | 0 | 3 |
| GATM | BRK1 | 0.729494 | 7 | 0 | 7 |
| GATM | FBXL3 | 0.726199 | 3 | 0 | 3 |
| GATM | PLEKHA7 | 0.717805 | 7 | 0 | 7 |
| GATM | SNORA68 | 0.714978 | 4 | 0 | 4 |
| GATM | SLC10A5 | 0.71457 | 3 | 0 | 3 |
| GATM | VSIG2 | 0.714161 | 5 | 0 | 5 |
| GATM | FAM3B | 0.712582 | 6 | 0 | 5 |
| GATM | FCHO2 | 0.709754 | 7 | 0 | 7 |
| GATM | GPD1L | 0.70908 | 11 | 0 | 10 |
| GATM | NAAA | 0.702984 | 8 | 0 | 8 |
| GATM | NDUFC1 | 0.699456 | 7 | 0 | 6 |
| GATM | FRMD6 | 0.695168 | 3 | 0 | 3 |
| GATM | C5orf66-AS1 | 0.694965 | 6 | 0 | 6 |
| GATM | MAML3 | 0.694087 | 4 | 0 | 3 |
| GATM | SFTA2 | 0.693886 | 4 | 0 | 4 |
| GATM | AK3 | 0.693744 | 7 | 0 | 6 |
| GATM | CASC3 | 0.692479 | 3 | 0 | 3 |
For details and further investigation, click here