| Full name: G-patch domain containing 4 | Alias Symbol: FLJ20249|DKFZP434F1735 | ||
| Type: protein-coding gene | Cytoband: 1q22-q23.1 | ||
| Entrez ID: 54865 | HGNC ID: HGNC:25982 | Ensembl Gene: ENSG00000160818 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of GPATCH4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GPATCH4 | 54865 | 224632_at | 0.7931 | 0.0997 | |
| GSE20347 | GPATCH4 | 54865 | 220596_at | 0.1196 | 0.1945 | |
| GSE23400 | GPATCH4 | 54865 | 221733_s_at | -0.2223 | 0.0000 | |
| GSE26886 | GPATCH4 | 54865 | 224634_at | 0.9865 | 0.0011 | |
| GSE29001 | GPATCH4 | 54865 | 221733_s_at | 0.0211 | 0.8939 | |
| GSE38129 | GPATCH4 | 54865 | 220596_at | 0.0297 | 0.7540 | |
| GSE45670 | GPATCH4 | 54865 | 224634_at | 0.6057 | 0.0000 | |
| GSE53622 | GPATCH4 | 54865 | 30035 | 0.3202 | 0.0000 | |
| GSE53624 | GPATCH4 | 54865 | 30035 | 0.5285 | 0.0000 | |
| GSE63941 | GPATCH4 | 54865 | 224634_at | 0.2167 | 0.6637 | |
| GSE77861 | GPATCH4 | 54865 | 224634_at | 0.6641 | 0.0355 | |
| GSE97050 | GPATCH4 | 54865 | A_33_P3211263 | 0.2796 | 0.4653 | |
| SRP007169 | GPATCH4 | 54865 | RNAseq | 2.4237 | 0.0008 | |
| SRP064894 | GPATCH4 | 54865 | RNAseq | 0.3033 | 0.1188 | |
| SRP133303 | GPATCH4 | 54865 | RNAseq | 0.6589 | 0.0003 | |
| SRP159526 | GPATCH4 | 54865 | RNAseq | 0.6921 | 0.0296 | |
| SRP193095 | GPATCH4 | 54865 | RNAseq | 0.4370 | 0.0005 | |
| SRP219564 | GPATCH4 | 54865 | RNAseq | 0.4065 | 0.2500 | |
| TCGA | GPATCH4 | 54865 | RNAseq | 0.0672 | 0.2014 |
Upregulated datasets: 1; Downregulated datasets: 0.
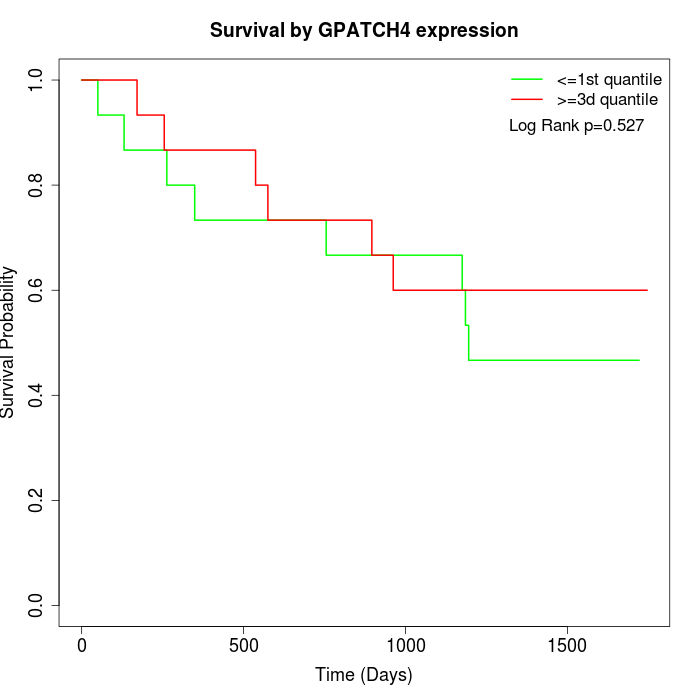
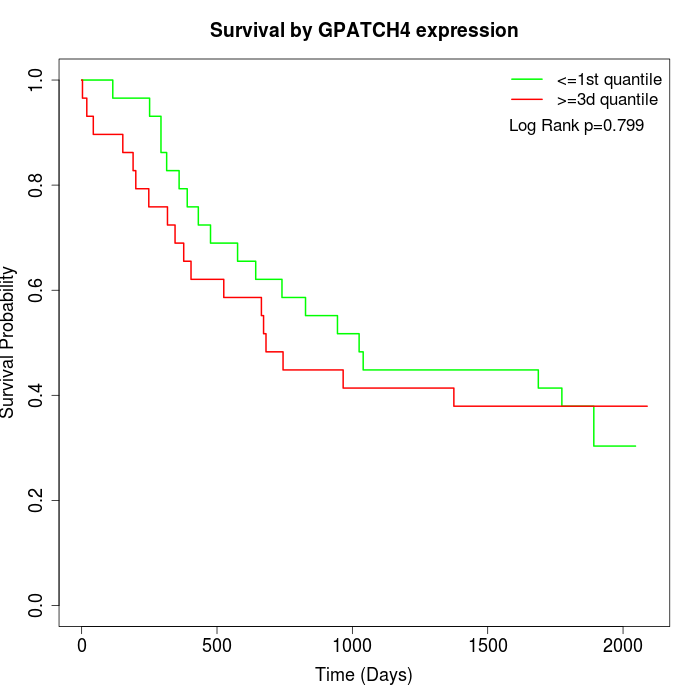
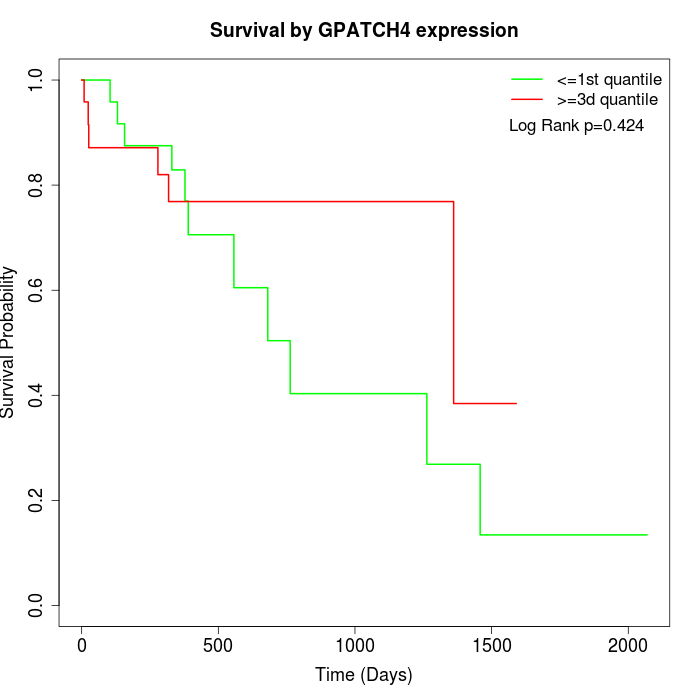
Survival by GPATCH4 expression:
Note: Click image to view full size file.
Copy number change of GPATCH4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GPATCH4 | 54865 | 12 | 0 | 18 | |
| GSE20123 | GPATCH4 | 54865 | 12 | 0 | 18 | |
| GSE43470 | GPATCH4 | 54865 | 7 | 2 | 34 | |
| GSE46452 | GPATCH4 | 54865 | 2 | 1 | 56 | |
| GSE47630 | GPATCH4 | 54865 | 14 | 0 | 26 | |
| GSE54993 | GPATCH4 | 54865 | 0 | 5 | 65 | |
| GSE54994 | GPATCH4 | 54865 | 16 | 0 | 37 | |
| GSE60625 | GPATCH4 | 54865 | 0 | 0 | 11 | |
| GSE74703 | GPATCH4 | 54865 | 7 | 2 | 27 | |
| GSE74704 | GPATCH4 | 54865 | 6 | 0 | 14 | |
| TCGA | GPATCH4 | 54865 | 38 | 2 | 56 |
Total number of gains: 114; Total number of losses: 12; Total Number of normals: 362.
Somatic mutations of GPATCH4:
Generating mutation plots.
Highly correlated genes for GPATCH4:
Showing top 20/886 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GPATCH4 | CRCP | 0.749593 | 3 | 0 | 3 |
| GPATCH4 | ZNF680 | 0.735213 | 3 | 0 | 3 |
| GPATCH4 | MDM4 | 0.73119 | 3 | 0 | 3 |
| GPATCH4 | CEP131 | 0.722109 | 4 | 0 | 4 |
| GPATCH4 | ZFP41 | 0.721562 | 4 | 0 | 4 |
| GPATCH4 | COG2 | 0.717944 | 6 | 0 | 6 |
| GPATCH4 | LSG1 | 0.717648 | 6 | 0 | 5 |
| GPATCH4 | COMMD5 | 0.713498 | 3 | 0 | 3 |
| GPATCH4 | KNOP1 | 0.712162 | 3 | 0 | 3 |
| GPATCH4 | FASN | 0.707452 | 3 | 0 | 3 |
| GPATCH4 | STAP1 | 0.700593 | 3 | 0 | 3 |
| GPATCH4 | E2F3 | 0.698429 | 7 | 0 | 7 |
| GPATCH4 | ZNF300 | 0.693093 | 3 | 0 | 3 |
| GPATCH4 | SAMD1 | 0.688397 | 3 | 0 | 3 |
| GPATCH4 | NUDCD2 | 0.688066 | 3 | 0 | 3 |
| GPATCH4 | TNFSF8 | 0.687349 | 4 | 0 | 4 |
| GPATCH4 | PNPT1 | 0.686084 | 8 | 0 | 7 |
| GPATCH4 | DENND6B | 0.682771 | 4 | 0 | 4 |
| GPATCH4 | ATAD3A | 0.682391 | 4 | 0 | 4 |
| GPATCH4 | PACS1 | 0.682208 | 3 | 0 | 3 |
For details and further investigation, click here