| Full name: goosecoid homeobox | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 14q32.13 | ||
| Entrez ID: 145258 | HGNC ID: HGNC:4612 | Ensembl Gene: ENSG00000133937 | OMIM ID: 138890 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of GSC:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GSC | 145258 | 1552338_at | -0.0509 | 0.8964 | |
| GSE26886 | GSC | 145258 | 1552338_at | 0.3758 | 0.0037 | |
| GSE45670 | GSC | 145258 | 1552338_at | 0.1209 | 0.2449 | |
| GSE53622 | GSC | 145258 | 85994 | 0.8286 | 0.0000 | |
| GSE53624 | GSC | 145258 | 85994 | 1.2833 | 0.0000 | |
| GSE63941 | GSC | 145258 | 1552338_at | 0.2607 | 0.3465 | |
| GSE77861 | GSC | 145258 | 1552338_at | -0.0787 | 0.3587 | |
| TCGA | GSC | 145258 | RNAseq | 3.4166 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 0.
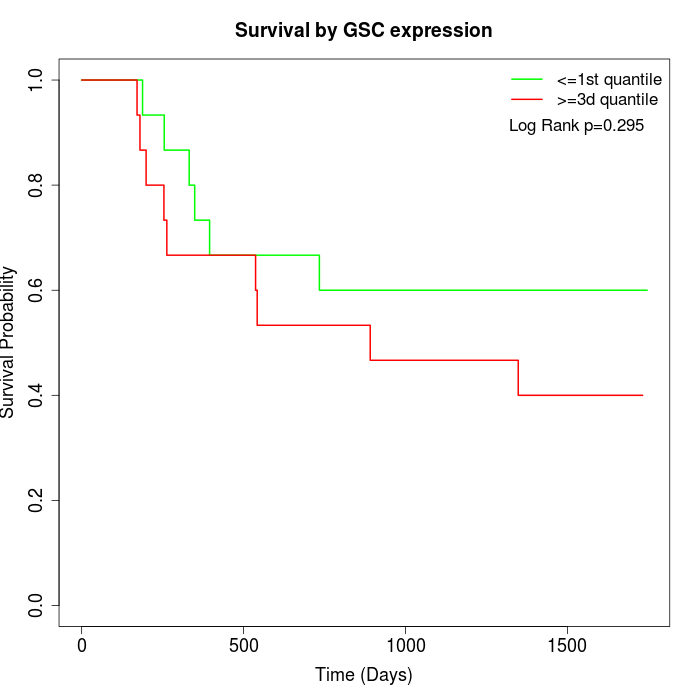
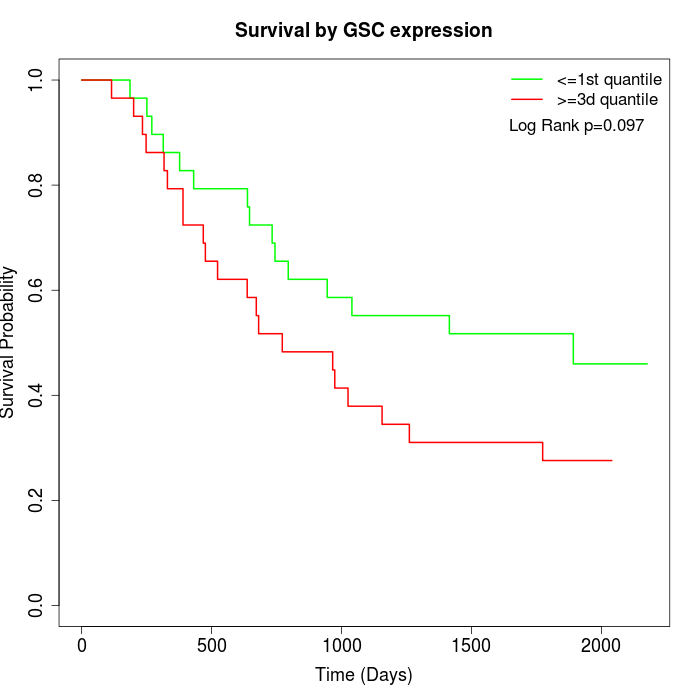
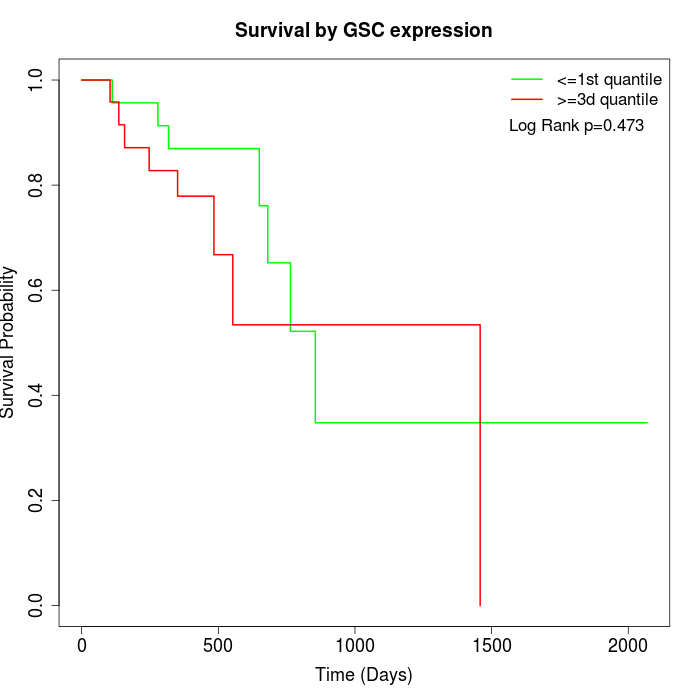
Survival by GSC expression:
Note: Click image to view full size file.
Copy number change of GSC:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GSC | 145258 | 10 | 4 | 16 | |
| GSE20123 | GSC | 145258 | 10 | 3 | 17 | |
| GSE43470 | GSC | 145258 | 8 | 3 | 32 | |
| GSE46452 | GSC | 145258 | 16 | 3 | 40 | |
| GSE47630 | GSC | 145258 | 11 | 8 | 21 | |
| GSE54993 | GSC | 145258 | 3 | 8 | 59 | |
| GSE54994 | GSC | 145258 | 21 | 4 | 28 | |
| GSE60625 | GSC | 145258 | 0 | 2 | 9 | |
| GSE74703 | GSC | 145258 | 6 | 3 | 27 | |
| GSE74704 | GSC | 145258 | 6 | 3 | 11 | |
| TCGA | GSC | 145258 | 30 | 20 | 46 |
Total number of gains: 121; Total number of losses: 61; Total Number of normals: 306.
Somatic mutations of GSC:
Generating mutation plots.
Highly correlated genes for GSC:
Showing top 20/69 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GSC | PPP1R1A | 0.702818 | 3 | 0 | 3 |
| GSC | SIX6 | 0.698996 | 3 | 0 | 3 |
| GSC | MUC2 | 0.677456 | 3 | 0 | 3 |
| GSC | ASB11 | 0.676757 | 3 | 0 | 3 |
| GSC | RPPH1 | 0.656872 | 3 | 0 | 3 |
| GSC | MCHR2-AS1 | 0.642279 | 3 | 0 | 3 |
| GSC | CDKN2B-AS1 | 0.624263 | 3 | 0 | 3 |
| GSC | KAZALD1 | 0.624004 | 3 | 0 | 3 |
| GSC | HMX2 | 0.61888 | 4 | 0 | 4 |
| GSC | SFMBT2 | 0.614363 | 3 | 0 | 3 |
| GSC | DLK1 | 0.605771 | 3 | 0 | 3 |
| GSC | LRRC3 | 0.601085 | 4 | 0 | 3 |
| GSC | DKKL1 | 0.600377 | 3 | 0 | 3 |
| GSC | ATP6V0E2-AS1 | 0.600359 | 3 | 0 | 3 |
| GSC | FOXL1 | 0.598001 | 4 | 0 | 4 |
| GSC | CHD5 | 0.597637 | 3 | 0 | 3 |
| GSC | CHST8 | 0.596725 | 3 | 0 | 3 |
| GSC | LINC00607 | 0.594041 | 3 | 0 | 3 |
| GSC | MIOX | 0.593403 | 3 | 0 | 3 |
| GSC | LRFN1 | 0.590334 | 3 | 0 | 3 |
For details and further investigation, click here