| Full name: goosecoid homeobox 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 22q11.21 | ||
| Entrez ID: 2928 | HGNC ID: HGNC:4613 | Ensembl Gene: ENSG00000063515 | OMIM ID: 601845 |
| Drug and gene relationship at DGIdb | |||
Expression of GSC2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GSC2 | 2928 | 221441_at | 0.0891 | 0.6964 | |
| GSE20347 | GSC2 | 2928 | 221441_at | -0.0169 | 0.7513 | |
| GSE23400 | GSC2 | 2928 | 221441_at | -0.0593 | 0.0040 | |
| GSE26886 | GSC2 | 2928 | 221441_at | 0.0754 | 0.5555 | |
| GSE29001 | GSC2 | 2928 | 221441_at | -0.0389 | 0.7135 | |
| GSE38129 | GSC2 | 2928 | 221441_at | -0.0406 | 0.3069 | |
| GSE45670 | GSC2 | 2928 | 221441_at | -0.0680 | 0.3563 | |
| GSE53622 | GSC2 | 2928 | 25860 | 0.1089 | 0.1134 | |
| GSE53624 | GSC2 | 2928 | 25860 | 0.1013 | 0.0120 | |
| GSE63941 | GSC2 | 2928 | 221441_at | 0.1691 | 0.2027 | |
| GSE77861 | GSC2 | 2928 | 221441_at | 0.0110 | 0.9219 | |
| GSE97050 | GSC2 | 2928 | A_23_P109382 | 0.1493 | 0.4892 |
Upregulated datasets: 0; Downregulated datasets: 0.
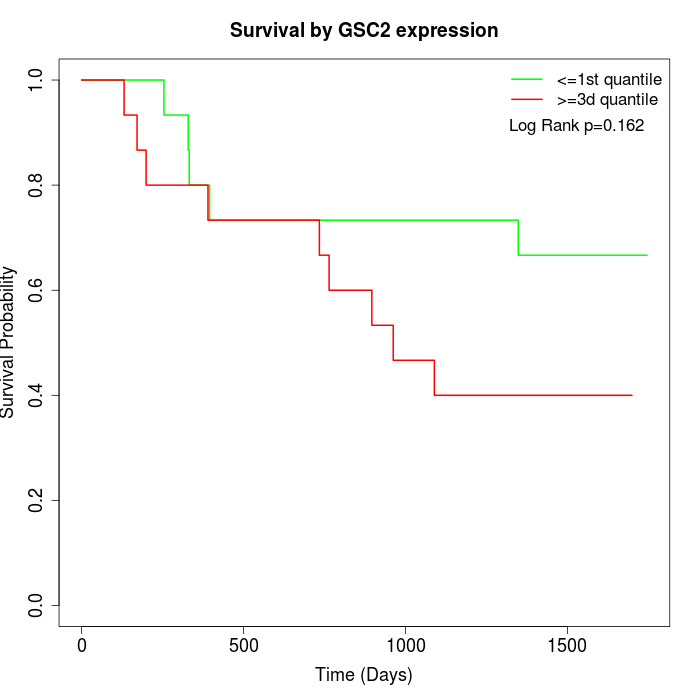
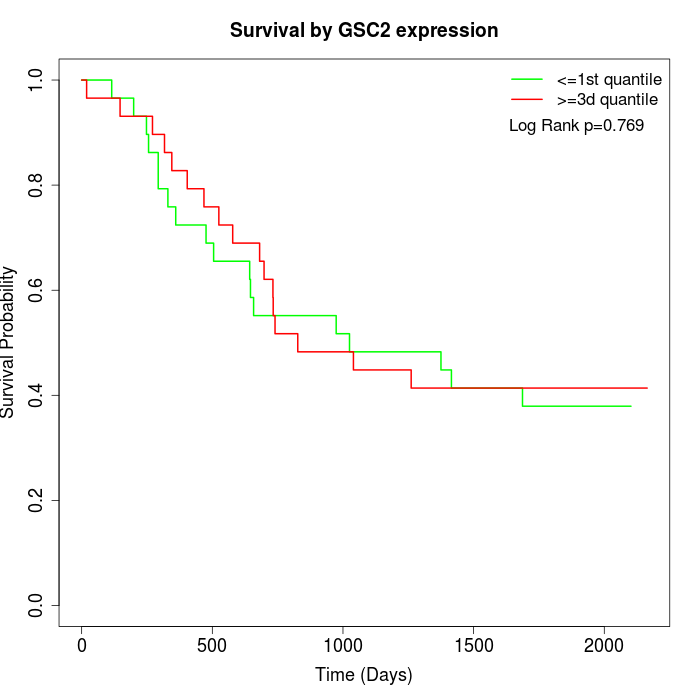
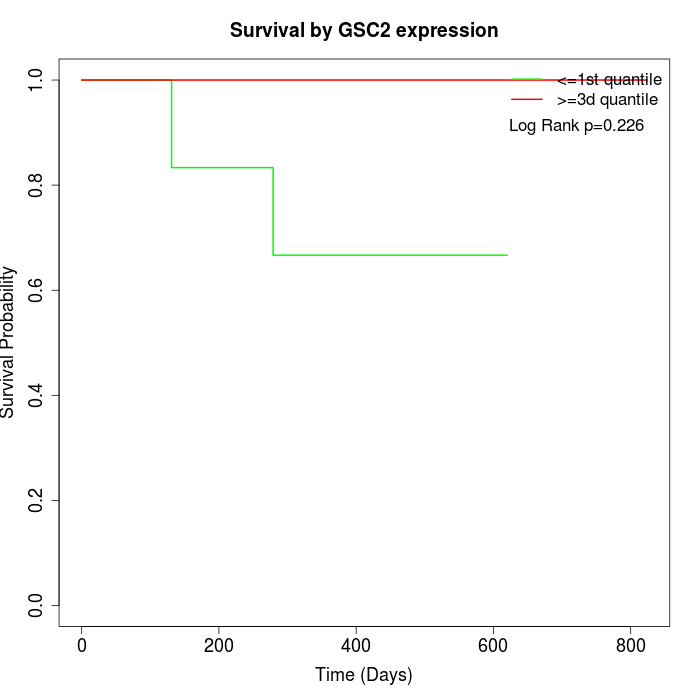
Survival by GSC2 expression:
Note: Click image to view full size file.
Copy number change of GSC2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GSC2 | 2928 | 4 | 8 | 18 | |
| GSE20123 | GSC2 | 2928 | 4 | 8 | 18 | |
| GSE43470 | GSC2 | 2928 | 5 | 7 | 31 | |
| GSE46452 | GSC2 | 2928 | 33 | 1 | 25 | |
| GSE47630 | GSC2 | 2928 | 8 | 5 | 27 | |
| GSE54993 | GSC2 | 2928 | 3 | 6 | 61 | |
| GSE54994 | GSC2 | 2928 | 12 | 7 | 34 | |
| GSE60625 | GSC2 | 2928 | 5 | 0 | 6 | |
| GSE74703 | GSC2 | 2928 | 5 | 5 | 26 | |
| GSE74704 | GSC2 | 2928 | 2 | 5 | 13 | |
| TCGA | GSC2 | 2928 | 27 | 17 | 52 |
Total number of gains: 108; Total number of losses: 69; Total Number of normals: 311.
Somatic mutations of GSC2:
Generating mutation plots.
Highly correlated genes for GSC2:
Showing top 20/140 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GSC2 | FBXO39 | 0.705238 | 3 | 0 | 3 |
| GSC2 | RGS3 | 0.703979 | 3 | 0 | 3 |
| GSC2 | RIT2 | 0.694389 | 3 | 0 | 3 |
| GSC2 | CNTF | 0.681225 | 3 | 0 | 3 |
| GSC2 | CLDN18 | 0.677691 | 3 | 0 | 3 |
| GSC2 | TNFRSF4 | 0.67641 | 4 | 0 | 3 |
| GSC2 | GRIN2D | 0.6741 | 3 | 0 | 3 |
| GSC2 | GNG3 | 0.664706 | 4 | 0 | 3 |
| GSC2 | TGFB2 | 0.663675 | 3 | 0 | 3 |
| GSC2 | ADAMTS10 | 0.663341 | 3 | 0 | 3 |
| GSC2 | ADCY3 | 0.655616 | 3 | 0 | 3 |
| GSC2 | TCP11L1 | 0.653291 | 3 | 0 | 3 |
| GSC2 | DOT1L | 0.65289 | 4 | 0 | 3 |
| GSC2 | PEAK1 | 0.652506 | 4 | 0 | 4 |
| GSC2 | ZNF467 | 0.649033 | 4 | 0 | 3 |
| GSC2 | TGFB3 | 0.645437 | 3 | 0 | 3 |
| GSC2 | SLITRK3 | 0.642335 | 3 | 0 | 3 |
| GSC2 | PCGF2 | 0.640858 | 3 | 0 | 3 |
| GSC2 | CCNK | 0.637732 | 4 | 0 | 4 |
| GSC2 | UTS2R | 0.636803 | 3 | 0 | 3 |
For details and further investigation, click here