| Full name: glycogen synthase 1 | Alias Symbol: GSY | ||
| Type: protein-coding gene | Cytoband: 19q13.33 | ||
| Entrez ID: 2997 | HGNC ID: HGNC:4706 | Ensembl Gene: ENSG00000104812 | OMIM ID: 138570 |
| Drug and gene relationship at DGIdb | |||
GYS1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04152 | AMPK signaling pathway | |
| hsa04910 | Insulin signaling pathway | |
| hsa04922 | Glucagon signaling pathway | |
| hsa04931 | Insulin resistance |
Expression of GYS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GYS1 | 2997 | 201673_s_at | 0.3201 | 0.4465 | |
| GSE20347 | GYS1 | 2997 | 201673_s_at | -0.1273 | 0.4386 | |
| GSE23400 | GYS1 | 2997 | 201673_s_at | 0.1535 | 0.0413 | |
| GSE26886 | GYS1 | 2997 | 201673_s_at | -0.1959 | 0.3062 | |
| GSE29001 | GYS1 | 2997 | 201673_s_at | -0.1214 | 0.6048 | |
| GSE38129 | GYS1 | 2997 | 201673_s_at | -0.0560 | 0.7065 | |
| GSE45670 | GYS1 | 2997 | 201673_s_at | 0.3404 | 0.0482 | |
| GSE53622 | GYS1 | 2997 | 36461 | -0.0902 | 0.2592 | |
| GSE53624 | GYS1 | 2997 | 36461 | 0.0661 | 0.2545 | |
| GSE63941 | GYS1 | 2997 | 201673_s_at | -0.0788 | 0.8729 | |
| GSE77861 | GYS1 | 2997 | 201673_s_at | 0.2113 | 0.4123 | |
| GSE97050 | GYS1 | 2997 | A_23_P208698 | -0.3354 | 0.2151 | |
| SRP007169 | GYS1 | 2997 | RNAseq | -0.1136 | 0.7615 | |
| SRP008496 | GYS1 | 2997 | RNAseq | -0.5150 | 0.0566 | |
| SRP064894 | GYS1 | 2997 | RNAseq | 0.4786 | 0.0122 | |
| SRP133303 | GYS1 | 2997 | RNAseq | -0.0091 | 0.9551 | |
| SRP159526 | GYS1 | 2997 | RNAseq | -0.0463 | 0.8502 | |
| SRP193095 | GYS1 | 2997 | RNAseq | 0.1688 | 0.1539 | |
| SRP219564 | GYS1 | 2997 | RNAseq | -0.3218 | 0.1374 | |
| TCGA | GYS1 | 2997 | RNAseq | 0.0446 | 0.3540 |
Upregulated datasets: 0; Downregulated datasets: 0.
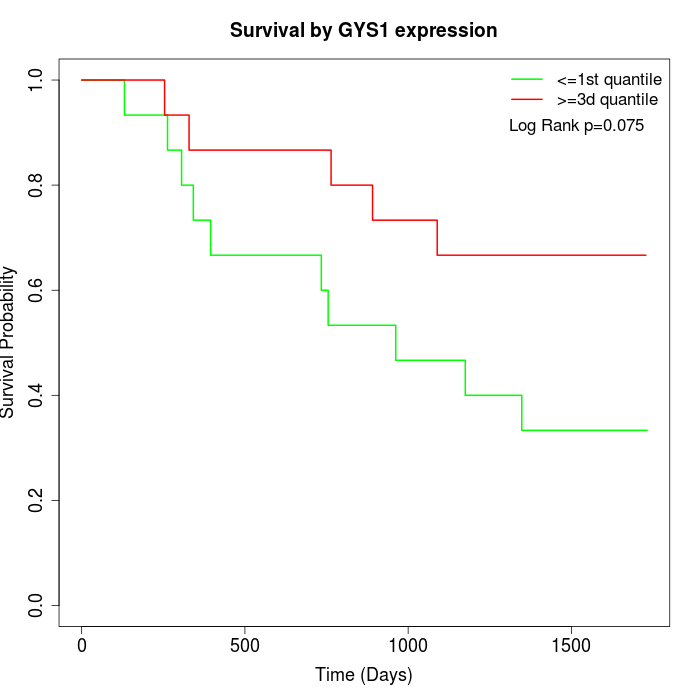
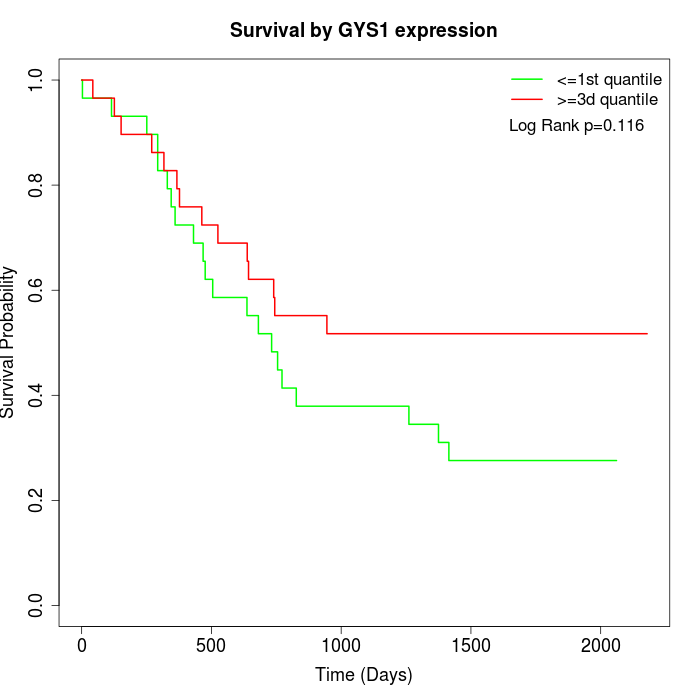
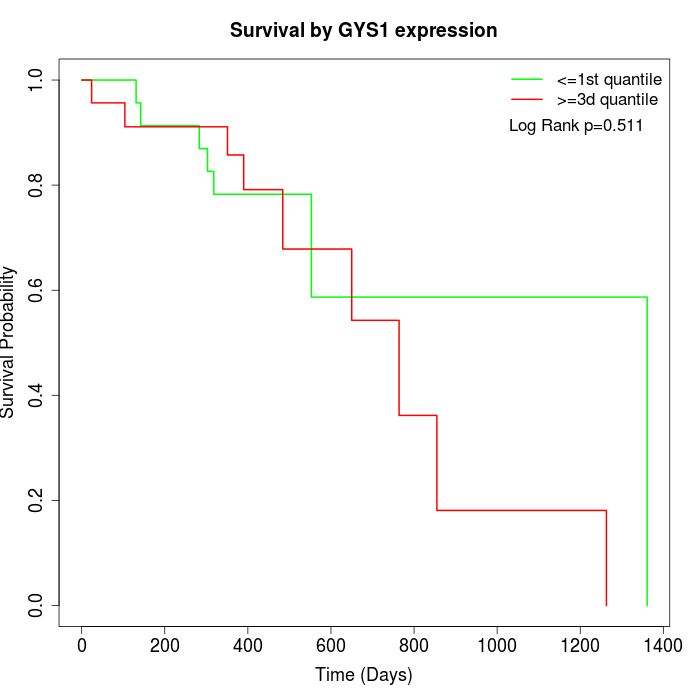
Survival by GYS1 expression:
Note: Click image to view full size file.
Copy number change of GYS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GYS1 | 2997 | 4 | 4 | 22 | |
| GSE20123 | GYS1 | 2997 | 4 | 3 | 23 | |
| GSE43470 | GYS1 | 2997 | 4 | 10 | 29 | |
| GSE46452 | GYS1 | 2997 | 45 | 1 | 13 | |
| GSE47630 | GYS1 | 2997 | 9 | 6 | 25 | |
| GSE54993 | GYS1 | 2997 | 17 | 4 | 49 | |
| GSE54994 | GYS1 | 2997 | 4 | 14 | 35 | |
| GSE60625 | GYS1 | 2997 | 9 | 0 | 2 | |
| GSE74703 | GYS1 | 2997 | 4 | 7 | 25 | |
| GSE74704 | GYS1 | 2997 | 4 | 1 | 15 | |
| TCGA | GYS1 | 2997 | 14 | 18 | 64 |
Total number of gains: 118; Total number of losses: 68; Total Number of normals: 302.
Somatic mutations of GYS1:
Generating mutation plots.
Highly correlated genes for GYS1:
Showing top 20/286 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GYS1 | MLLT1 | 0.778256 | 3 | 0 | 3 |
| GYS1 | DMAP1 | 0.757317 | 3 | 0 | 3 |
| GYS1 | ZNF284 | 0.734426 | 3 | 0 | 3 |
| GYS1 | CACNA1B | 0.728445 | 3 | 0 | 3 |
| GYS1 | RPN1 | 0.727936 | 3 | 0 | 3 |
| GYS1 | DDX6 | 0.724969 | 3 | 0 | 3 |
| GYS1 | HDAC6 | 0.724226 | 3 | 0 | 3 |
| GYS1 | TFDP2 | 0.724002 | 3 | 0 | 3 |
| GYS1 | ZBTB11 | 0.715786 | 3 | 0 | 3 |
| GYS1 | BAG6 | 0.715528 | 4 | 0 | 3 |
| GYS1 | NCSTN | 0.715322 | 3 | 0 | 3 |
| GYS1 | KLHDC3 | 0.710204 | 3 | 0 | 3 |
| GYS1 | TRAM1 | 0.705411 | 3 | 0 | 3 |
| GYS1 | SLC45A4 | 0.705368 | 3 | 0 | 3 |
| GYS1 | TBCD | 0.703216 | 3 | 0 | 3 |
| GYS1 | PDRG1 | 0.701371 | 3 | 0 | 3 |
| GYS1 | NDUFB4 | 0.700524 | 3 | 0 | 3 |
| GYS1 | LEMD2 | 0.699969 | 4 | 0 | 3 |
| GYS1 | ARHGAP35 | 0.698295 | 4 | 0 | 3 |
| GYS1 | ZNF74 | 0.696029 | 3 | 0 | 3 |
For details and further investigation, click here