| Full name: nicastrin | Alias Symbol: KIAA0253|APH2 | ||
| Type: protein-coding gene | Cytoband: 1q23.2 | ||
| Entrez ID: 23385 | HGNC ID: HGNC:17091 | Ensembl Gene: ENSG00000162736 | OMIM ID: 605254 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
NCSTN involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04330 | Notch signaling pathway |
Expression of NCSTN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NCSTN | 23385 | 208759_at | 0.4714 | 0.1029 | |
| GSE20347 | NCSTN | 23385 | 208759_at | 0.2775 | 0.0019 | |
| GSE23400 | NCSTN | 23385 | 208759_at | 0.2855 | 0.0000 | |
| GSE26886 | NCSTN | 23385 | 208759_at | 0.1314 | 0.4793 | |
| GSE29001 | NCSTN | 23385 | 208759_at | 0.2933 | 0.2532 | |
| GSE38129 | NCSTN | 23385 | 208759_at | 0.3585 | 0.0002 | |
| GSE45670 | NCSTN | 23385 | 208759_at | 0.2036 | 0.0401 | |
| GSE53622 | NCSTN | 23385 | 1210 | 0.1872 | 0.0024 | |
| GSE53624 | NCSTN | 23385 | 33872 | -0.6681 | 0.0000 | |
| GSE63941 | NCSTN | 23385 | 208759_at | 0.0013 | 0.9982 | |
| GSE77861 | NCSTN | 23385 | 208759_at | 0.3469 | 0.2303 | |
| GSE97050 | NCSTN | 23385 | A_23_P34402 | -0.2442 | 0.2882 | |
| SRP007169 | NCSTN | 23385 | RNAseq | 0.1284 | 0.7095 | |
| SRP008496 | NCSTN | 23385 | RNAseq | 0.2180 | 0.3688 | |
| SRP064894 | NCSTN | 23385 | RNAseq | 0.4370 | 0.0041 | |
| SRP133303 | NCSTN | 23385 | RNAseq | 0.3014 | 0.0059 | |
| SRP159526 | NCSTN | 23385 | RNAseq | 0.5351 | 0.0064 | |
| SRP193095 | NCSTN | 23385 | RNAseq | 0.2566 | 0.0173 | |
| SRP219564 | NCSTN | 23385 | RNAseq | 0.4431 | 0.0983 | |
| TCGA | NCSTN | 23385 | RNAseq | 0.0367 | 0.3752 |
Upregulated datasets: 0; Downregulated datasets: 0.
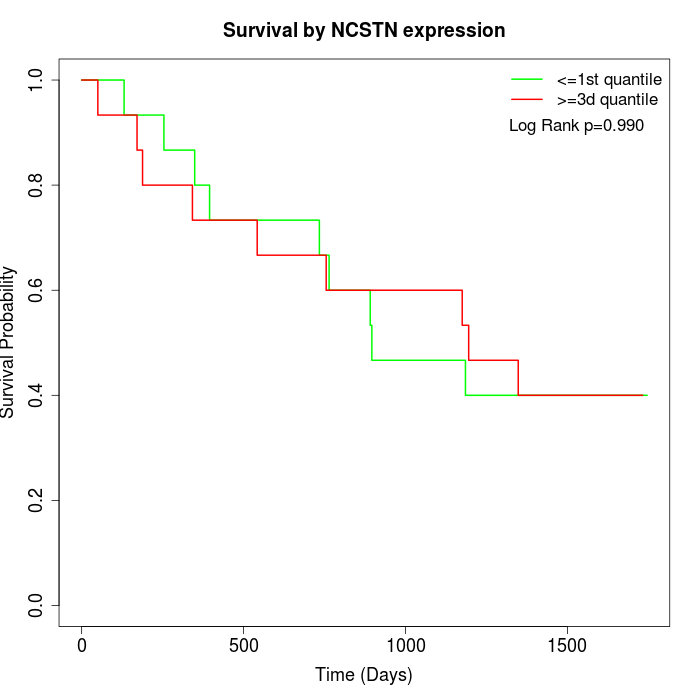
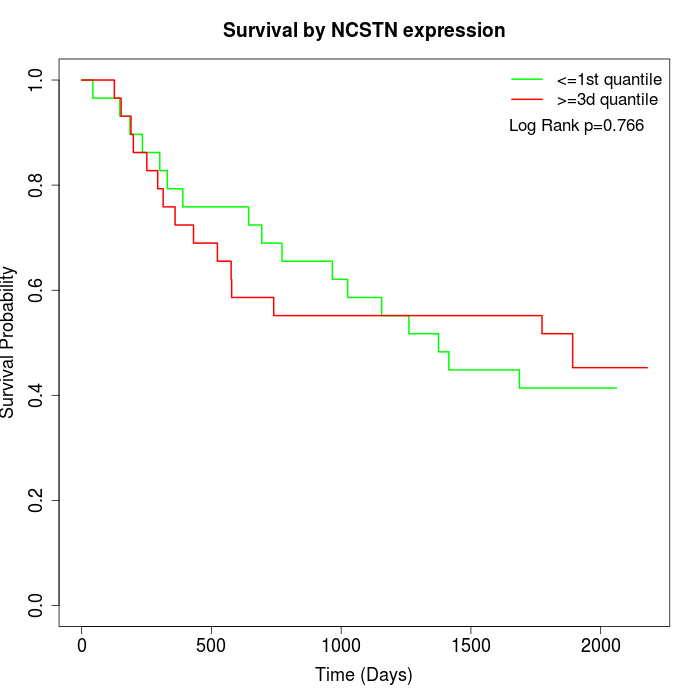
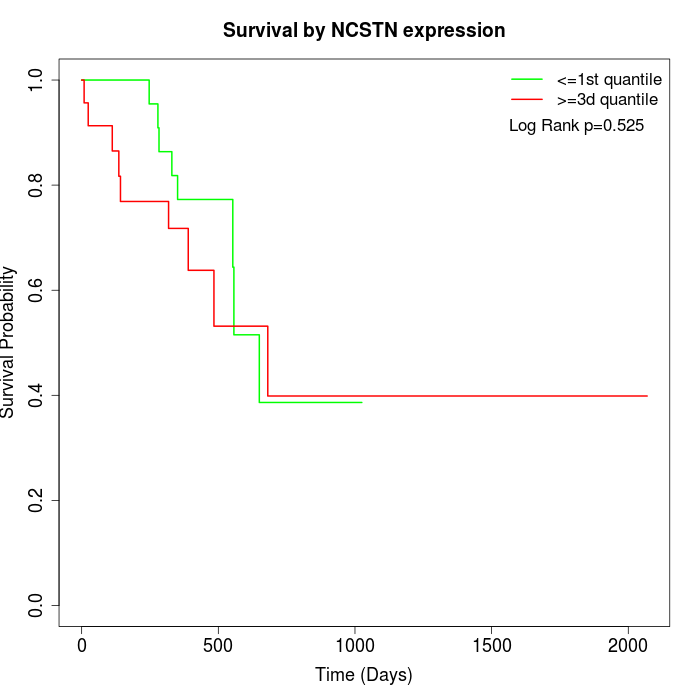
Survival by NCSTN expression:
Note: Click image to view full size file.
Copy number change of NCSTN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NCSTN | 23385 | 10 | 0 | 20 | |
| GSE20123 | NCSTN | 23385 | 10 | 0 | 20 | |
| GSE43470 | NCSTN | 23385 | 7 | 2 | 34 | |
| GSE46452 | NCSTN | 23385 | 2 | 1 | 56 | |
| GSE47630 | NCSTN | 23385 | 14 | 0 | 26 | |
| GSE54993 | NCSTN | 23385 | 0 | 5 | 65 | |
| GSE54994 | NCSTN | 23385 | 16 | 0 | 37 | |
| GSE60625 | NCSTN | 23385 | 0 | 0 | 11 | |
| GSE74703 | NCSTN | 23385 | 7 | 2 | 27 | |
| GSE74704 | NCSTN | 23385 | 4 | 0 | 16 | |
| TCGA | NCSTN | 23385 | 44 | 3 | 49 |
Total number of gains: 114; Total number of losses: 13; Total Number of normals: 361.
Somatic mutations of NCSTN:
Generating mutation plots.
Highly correlated genes for NCSTN:
Showing top 20/323 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NCSTN | ZNF284 | 0.782851 | 3 | 0 | 3 |
| NCSTN | TNFRSF1A | 0.781942 | 3 | 0 | 3 |
| NCSTN | IRF2 | 0.761611 | 3 | 0 | 3 |
| NCSTN | DGCR2 | 0.729263 | 5 | 0 | 4 |
| NCSTN | GATAD2B | 0.729132 | 4 | 0 | 3 |
| NCSTN | RRM2B | 0.727282 | 3 | 0 | 3 |
| NCSTN | TAF4B | 0.717741 | 3 | 0 | 3 |
| NCSTN | GYS1 | 0.715322 | 3 | 0 | 3 |
| NCSTN | ZBED1 | 0.714022 | 3 | 0 | 3 |
| NCSTN | STRADA | 0.710646 | 4 | 0 | 3 |
| NCSTN | KDM1B | 0.708512 | 3 | 0 | 3 |
| NCSTN | RALGAPA2 | 0.706297 | 3 | 0 | 3 |
| NCSTN | CHD3 | 0.703688 | 6 | 0 | 6 |
| NCSTN | IFT80 | 0.699593 | 3 | 0 | 3 |
| NCSTN | MDM2 | 0.695777 | 3 | 0 | 3 |
| NCSTN | TERF1 | 0.695157 | 3 | 0 | 3 |
| NCSTN | DNM3 | 0.68992 | 3 | 0 | 3 |
| NCSTN | KRTCAP2 | 0.688704 | 4 | 0 | 3 |
| NCSTN | NOM1 | 0.688431 | 3 | 0 | 3 |
| NCSTN | ZNF496 | 0.687026 | 3 | 0 | 3 |
For details and further investigation, click here